In Situ Atomic Force Microscopy Studies of the Effect of Indolicidin on E.coli Cells
Hans Jakob Askou, Rasmus Neergaard Jakobsen and Peter Fojan*
Department of Physics and Nanotechnology, Aalborg University, Skjernvej 4A, 9220 Aalborg, Denmark
E-mail: fp@nano.aau.dk
∗Corresponding Author
Received 02 August 2018; Accepted 29 November 2018;
Publication 24 December 2018
Abstract
E.coli cells were succesfully attached to both gelatin coated surfaces and polylactic acid honeycomb patterned mica surfaces as determined by in situ atomic force microscopy. The gelatin coated surfaces provided a softer support onto which the E.coli cells were capable of slightly submerging leading to a better adhesion compared to the harder surfaces consisting of polylactic acid polymer surfaces. After continuous scanning in liquid media, the E.coli cells remained rod shaped and smooth. Indolicidin, a 13-AA linear antimicrobial peptide, was injected in order to visualize the peptide-membrane interactions in real time. Instantly after the injection of the peptides, the bacterial membranes were observed to be distorted and seemed to melt proceeding as a function of time. In conclusion, these experiments proved that the E.coli cells were not ruptured as could be expected due to pore formation and disruption of the osmotic pressure. This indicates a possible intracellular target killing mechanism of indolicidin interacting with E.coli cells.
Keywords
- Atomic force microscopy
- indolicidin
- antimicrobial peptides
- E.coli cells
- gelatin
- honeycomb pattern
- PLA
- intracellular killing mechanism
1 Introduction
Light microscopy has been used extensively for identification and quantification of microorganisms. However, the resolution of optical microscopy is limited by the wavelength of the applied light source, hence information at the nanometer level is not accessible with this method [1]. High resolution images of microbial samples can be obtained using electron microscopy, which uses high energy electrons instead of light as the incident beam. Although high-resolution imaging of bacteria is possible with electron microscopy, cells must be fixed to the substrate with cross-linking agents and imaged under vacuum. These conditions prevent imaging of living cells. Several reports have dealt with imaging of individual microbial cells after chemical fixation and/or dehydration. These pretreatments improve the imaging resolution, however, the surface of the microorganisms will not entirely be representative for the hydrated surface [2]. Near-field microscopes, such as scanning probe microscopes (SPM), utilize a small probe in close proximity to the surface of the sample and, by scanning the probe across the surface, a three-dimensional image of the surface topography is obtained [3, 4].
Atomic force microscopy (AFM) is a scanning probe technique, which has proven to be a useful technique to elucidate surface structure of biological materials at unprecedented resolution. Improvements of the imaging in liquid using AFM have enabled operation in aqueous solutions and under physiological conditions, which makes it possible to monitor important biological processes in real time at high resolution. Until now, AFM has been widely used to investigate the morphology of both prokaryotic and eukaryotic cells and also to measure interaction forces at biological interfaces. Among the studies dealing with entire bacterial cells, most of them were morphological studies performed in air, which limits the relevance of the results [5–7]. Imaging in liquid conditions that are compatible with the natural environment of living systems will be necessary for determining the function and dynamics of these systems [6, 7].
Phase imaging is a powerful extension of tapping mode AFM that provides additional nanometer-scale information about surface structure [8]. As the cantilever is excited into resonance oscillation with a piezoelectric driver, the oscillation amplitude is used as a feedback signal to measure topographic variations of the sample. In phase imaging, the phase lag of the cantilever oscillation, relative to the signal sent to the cantilever’s piezo driver, is simultaneously monitored. By mapping the phase of the cantilever oscillation, phase imaging can give detailed information about variations in composition, adhesion, friction, and viscoelasticity. Applications include identification of contaminants, mapping of different components in composite materials, and differentiating regions of high and low surface adhesion or hardness. Furthermore, phase imaging can act as a real-time contrast enhancement technique. Because phase imaging highlights edges and is not affected by large-scale height differences, it provides for clearer observation of fine features, such as grain edges, which can be obscured by rough topography [9].
In order to image a biological sample using AFM, the sample must be attached to a solid, flat substrate. Mica is widely used as a substrate in fields such as scanning probe microscopy, biotechnology, and material science, because its perfect cleavage, atomically flat and chemically inert surfaces makes it suitable for depositing biological materials, thin metallic films, or ordered molecular layers [10]. Immobilization of bacteria on a surface is a complicated process that is influenced by many factors including the surface energy and charge of the bacteria as well as the substrate, the chemical composition, roughness of the substrate, and environmental factors such as temperature and time of exposure [11]. In general, surface roughness leads to improved bacterial adhesion [12, 13]. It has been reported that bacteria preferentially attach to micro-scale surface features such as scratches, pits, and grooves (e.g. irregularities of polymeric surfaces) [14, 15]. Furthermore, living bacteria often have flagella allowing them to move through the growth medium and, if they choose to attach to an AFM substrate, they can frequently detach and swim away. If they choose to remain, they often synthesize extrapolymeric substances, such as alginate, which obscur cell walls. If cells are not motile, there will be a reliance on Brownian motion to (eventually) bring them into contact with the AFM substrate. Once contact has been established, the initial adhesion forces are reversible and weak so that lateral tip forces exerted by an AFM cantilever can be sufficient to dislodge the cells [16].
Although several studies regarding attachment and visualization of bacteria have been performed, not many have actually visualized bacteria using in situ AFM (here, in situ referes to real time operation in liquid). The most pronounced techniques for bacterial attachment are modification of the substrate surface with gelatine [6], poly-L-lysine [8], other organic polymers [17] or attachment to a membrane surface [7]. Gelatin coated mica surfaces have been shown to be suitable for immobilizing and imaging both gram positive and gram negative bacteria in air and liquid environments as well [5, 6]. Furthermore, a polymer technique using polylactic acid (PLA) and different micelles/vesicles as patterning agent has shown to create a PLA-honeycomb-patterned film which has been reported to be useful for scaffolds in cell culture tissue engineering [17, 18].
Along with the discovery of new diseases, bacterial resistance is becoming a growing threat in medicine. Over the past several decades, the search for new drugs and treatments has been focused on a group of short, 10–50-residue polypeptides, called antimicrobial peptides (AMPs) [19, 20]. AMPs are important components of the innate immune system of multicellular organisms to control the natural flora and combat a wide spectrum of pathogens. Considering the millions of years they have been used successfully in nature they are still highly effective today, as compared to classical antibiotics. AMPs are found in many different organisms ranging from fungi, plants and insects to animals, including crustaceans, birds, fish, and humans [21, 22]. Importantly, many AMPs rapidly permeate and destroy the cell membrane, causing severe damage. This is in contrast to conventional antibiotics, which act on specific targets such as enzymes or DNA replication. Furthermore, AMPs have a broad range of targets and perform their action on the entire cellular membrane, therefore it is believed that bacteria will only develop tolerance with great difficulty [22]. Thus, it has been reported that AMPs will be able to contribute as therapeutic agents [19, 20].
AMPs are typically amphipathic peptides and are often cationic at physiological pH showing high affinities towards bacterial membranes, which carry a net negative charge. In contrast, the outer leaflets of mammalian cell membranes are mainly composed of zwitterionic phospholipids leading towards a lowered affinity of the AMPs [19]. Furthermore, the acidic character of the outer leaflet of the prokaryotic membrane facilitates its interaction with the usually cationic AMPs. Another major difference in the chemical composition of membranes between prokaryotic and eukaryotic cells is that the latter is abundant in sterols. Finally, the large inside negative transmembrane potential of bacterial cells may also lead to the formation of ion channels [19]. There is a consensus, however, that one major step in the activity of cationic AMPs is their initial binding to the negatively charged lipopolysaccharide (LPS) of Gram-negative bacteria or to the lipoteichoic acid of Gram-positive bacteria [23].
Understanding the mechanism of membrane permeation is crucial for the developmentof AMPs as future antibiotics. Regarding bacteria, the lipidic bacterial membrane and the negatively charged bacterial surface are the major targets for most AMPs. However, to reach their target membrane, the AMPs have to transverse the bacterial wall, which is different in Gram-negative and Gram-positive bacteria. Gram-negative bacteria have a smaller cell wall peptidoglycan layer than Gram-positive bacteria. In addition, Gram-negative bacteria also contain an outer membrane in addition to the common cytoplasmic membrane. Therefore, AMPs need to pass through two lipidic membranes in Gram-negative bacteria compared to only one membrane in Gram-positive bacteria [24]. However, the two membranes in Gram-negative bacteria actually cause AMPs to work as well or even better against Gram-positive bacteria. This is due to a specific uptake pathway across the outer membrane of Gram-negative bacteria, known as self-promoted uptake [25].
Indolicidin (ILPWKWPWWPWRR-NH2), a 13 amino acid cationic AMP rich in Trp with an extended structure is found in the cytoplasmic granules of bovine neutrophils [26]. Indolicidin is one of the shortest known natural-occurring AMPs and is toxic to both prokaryotes and eukaryotes alike and acts as an inhibitor/modulator for various enzymes [27, 28]. Indolicidin has been shown to possess antimicrobial activity against different microorganisms including Gram-positive and Gram-negative bacteria, fungi, and protozoa. Indolicidin is reported to inhibit the macromolecular synthesis in bacteria, which is common for the peptide family. It has been demonstrated that indolicidin inhibits the DNA synthesis in E.coli by inducing filaments in the bacteria [29].
Several mechanisms of action have been proposed for indolicidin depending on AMP concentration and membrane composition. Some of these include (i) cytoplasmic membrane permeabilization by ion channel formation, (ii) transient trans-membrane aqueous pore formation, (iii) interfacial membrane insertion followed by peptide-lipid toroidal pore formation, (iv) head-to-tail dimerization across the bilayer, and (v) inhibition of DNA synthesis after self-promoted uptake into the cytoplasm [30–32]. The mode of action for indolicidin has previously been described by the Shai-Matsuzaki-Huang model which proposes a stepwise interaction of the peptide with the membrane. First, the outer leaflet is covered with peptide (carpet mechanism) and the peptide integrates into the membrane causing a thinning of the outer leaflet. The surface area of the outer leaflet expands relative to the inner leaflet, resulting in a strain within the bilayer followed by phase transition and ‘worm-hole’ formation. Now the peptides can access the inner leaflet and in some cases diffuse towards intracellular targets. Eventually, the cell will collapse due to physical disruption of the cell membrane [33].
Trp- and Arg-rich AMPs are important as these amino acids represent a recurring theme among many AMPs and play an important role in antimicrobial activity [34]. Trp has a distinct preference for the interfacial region of lipid bilayers, while Arg residues endow the peptides with cationic charges and hydrogen bonding properties necessary for interaction with the abundant anionic components of bacterial membranes such as LPS, teichoic acid, or phosphatidylglycerol head groups normally present at the bacterial surface. Hereafter, Trp enables prolonged association of the peptide with the membrane [34, 35]. In combination, these two residues are capable of participating in cation–π interactions, thereby facilitating enhanced peptide–membrane interactions [36].
The primary interaction of peptides with a high content of Trp residues, including indolicidin, will preferentially be at the membrane-solution interface as this is most energetically favourable. The insertion of the hydrophobic residues of these peptides into a membrane may produce an increase in the lateral pressure near the interface, causing a disruption of the lipid chain packing [37]. In a previous study, we have observed that indolicidin induced a continuous membrane shrinking and thinning of model membranes composed of supported planar bilayers (SPBs) consisting of zwitterionic phosphatidylcholine (PC), anionic phosphatidylglycerol (PG), cationic ethylphosphocholine (EPC). Furthermore, a bacterial membrane was mimicked using a phospholipid model membrane composed of PG:PC 1:4 as this lipid composition correponds to the membrane composition of a bacteria with regards to phospholipid composition [38].
In a previous study it has been shown that the mechanism of action of indolicidin could be best seen and studied at the boundary of the gel-fluid-domains when imaged by in situ AFM. The physical properties as well as head groups and overall charge of the lipids play a crucial role. The specificity of the peptide is dependent upon the composition of the membrane. The different phases of lipid membranes observed during AFM imaging suggest that the process of indolicidin-induced membrane thinning requires phase-segregation where indolicidin acts at the boundary of the fluid and gel domains. Furthermore, it is believed that indolicidin induces a phase transistion from gel-phase to fluid-phase in the phospholipid bilayers [38].
A better understanding of the nature of the interaction between indolicidin and bacterial membranes will be an important step towards directed AMP evolution as novel antibiotics and the action of AMPs in general. With appropriate knowledge about the target organism, it should be possible to engineer a peptide with charge properties complementary to the membrane, but not too strong to prevent aggregation, and with a hydrophobicity best suited to permeabilize the membrane.
In this paper, we present the immobilization of E.coli cells on coated mica surfaces, as these have been shown to be suitable for immobilization and imaging of both gram positive and gram negative bacteria in air and liquid environments [5, 6]. The adhering properties of E.coli to the coated mica surfaces were compared and the surfaces showed biocompatibility. This enabled the investigations of the interactions between indolicidin and E.coli cells using real time in situ AFM operating in tapping mode.
2 Experimental Procedures
2.1 Chemicals
Indolicidin > 95% was bought from Ross Petersen Aps. Hexadecyltrimethylammonium bromide (CTAB)≥99.0%, Iron(III) chloride anhydrous ≥ 98% and Chloroform 99.8+% were purchased from Sigma-Aldrich. Polylactic acid (PLA) Mw∼55,000 was purchased from Fluka. Gelatine 220/230 Kloom SG718-F was purchased from SFK, and E.coli DH5α, Life Technologies, Inc. All chemicals have been used as supplied without any further purification steps prior to use.
2.2 Atomic Force Microscopy Imaging
AFM images were acquired at ambient temperature on a Digital Instruments Nanoscope IIIa Multimode (Digital Instruments/Veeco, Santa Barbara, CA) equipped with a ‘J’ scanner having a maximum lateral scan area of 160 μm × 160 μm, using 115 μm long silicon nitride triangular cantilevers NP-S - NP series’ from Veeco, with a nominal spring constant of 0.58 N/m (provided by the manufacturer). The mica surfaces investigated were glued onto a Teflon wafer to keep the fluid in the liquid cell tip holder and on the samples as the O-ring was removed from the fluid cell to prevent friction problems between the scan head and the sample. A contact/tapping mode fluid cell was placed over the Teflon wafer with the attached sample. The fluid cell was equipped with inlet tubes to allow injections of fluid into the chamber between measurements without removing the fluid cell. All AFM images were acquired using tapping mode and images were captured at a resolution of 512 × 512 pixels. The scan rate was adjusted for each sample and was between 0.4 and 1 Hz.
2.3 Honeycomb-patterned Polymer Surface
The honeycomb-patterned polymer surface was prepared by dissolving PLA in chloroform to a final concentration of 5 mg/mL. 20 μL of 12.5 mg/mL CTAB in milli-Q water was added to 1 mL PLA solution and was subsequently vortexed for 2 minutes. A freshly cleaved mica surface, mounted on a Teflon wafer, was covered with PLA-CTAB solution and was allowed to evaporate for five minutes. The surface was dried under a stream of nitrogen.
2.4 Gelatin Coated Surface
The gelatine coated surface was prepared by dissolving 0.25 g gelatine and 5 mg iron(III) chloride in 50 mL milli-Q-water. The solution was heated to 90∘C and subsequently cooled to 60∘C. A freshly cleaved mica disc, glued onto a Teflon wafer, was submerged in the solution and was placed vertically in a petri dish and left for drying overnight.
2.5 Bacterial Attachment
E.coli (DH5α) was grown in Luria Broth (LB) medium overnight at 37∘C at 260 rpm and was transferred onto LB plates. The bacteria were grown for 5 hours at 37∘C on plates, were harvested and dissolved in milli-Q water (OD = 0.6). Honeycomb-patterned polymer surfaces were incubated with bacteria for 30 minutes, whereas gelatine coated surfaces were incubated with bacteria for 1 to 5 minutes. The surfaces were rinsed using centrifuged milli-Q water (10,000 rpm, 15∘C, 15 minutes). All samples were imaged immediately after preparation.
2.6 Addition of Antimicrobial Peptide
25 μM indolicidin was injected into the fluid cell to monitor the effect of the peptides interacting with the bacteria in real time. The peptide solution was injected directly into the fluid cell via cannula transfer once stable images of bacteria were obtained, as determined by in situ AFM imaging. Here, stability refers to bacteria remaining unaffected by the continuous scanning by the tip on the surface. To investigate the peptide-bacterial interactions as a function of time, continuous AFM imaging was performed as a solution of indolicidin was injected in the liquid cell. Approx. 100 μL peptide solution was injected at time intervals of approx. 2 hours due to evaporation of the liquid and formation of air bubbles in the fluid cell. Prior to the addition of peptide solution, stability images were recorded using 10 mM HEPES buffer pH 7.0. It is therefore important to notice that the first addition of peptide solution results in a dilution of the injected peptide solution; therefore the absolute concentration of peptides actually is below 25.0 μM.
2.7 Image Processing
All images were flattened and plane fitted using WSxM v4.0 develop 8.6 software [39]. Furthermore, images containing single disrupted lines were processed using the filter Remove Lines, which performs a linear interpolation between the closest two ‘good lines’ and the lines selected to be removed.
3 Results
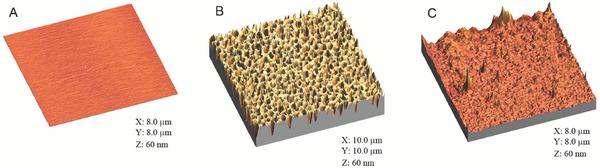
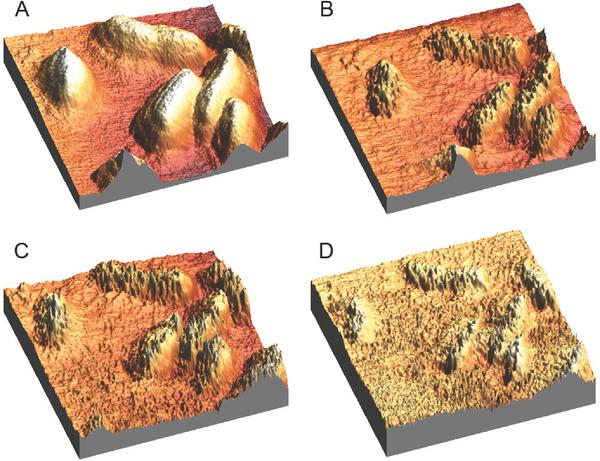
Two different surfaces were investigated in this study; (i) gelatin coated mica surfaces and (ii) polymer-coated mica surfaces. The overall surface roughness of the coated mica discs was investigated by AFM as this is an important factor in bacterial adhesion. AFM images and roughness analyses indicated micro- and nano structured surfaces on the gelatin coated and PLA-honeycomb polymer surfaces, whereas an untreated mica disc provides an almost atomically flat surface, see Figure 1 and Table 1. The main difference in topography between the polymer and gelatin surface, is the mesh of pores in the surface, created when applying the polymer whereas the gelatin surface appears more roughly structured, see Figures 1B and C, respectively. Pore sizes were between 300 to 600 nm in diameter and the measured values for surface heights, average heights, and RMS roughness are summarized in Table 1. From the roughness analysis it can be seen that the honeycomb-pattern provided the largest degree of surface roughness as expected.
Figure 1 3D AFM height images of surface roughness investigation of (A) a pure mica surface, (B) a PLA-honeycomb-patterned polymer surface, and (C) a gelatin coated surface. Scan area is listed with the images and height scale is 60 nm. All scans were performed in imaging buffer.
Table 1 Roughness analysis
| Surface | Max height/[nm] | Average height/[nm] | RMS roughness/[nm] |
| Mica | 2.50 | 0.61 | 0.12 |
| Honeycomb | 60.3 | 32.70 | 7.46 |
| Gelatin | 66.9 | 16.89 | 2.71 |
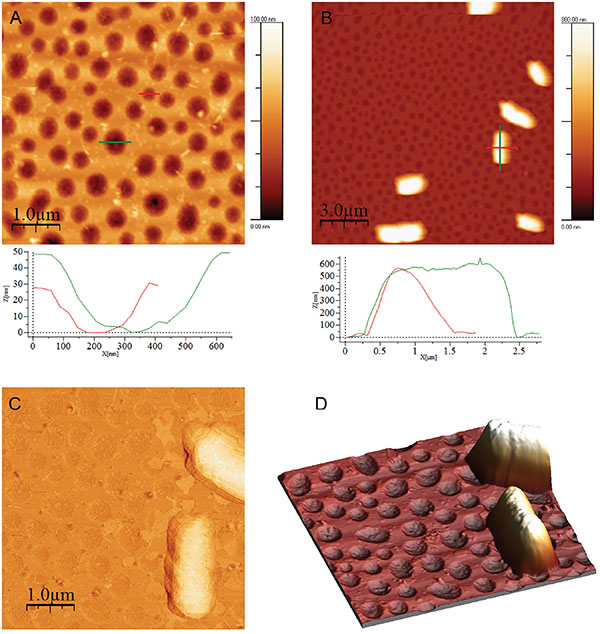
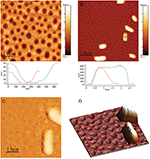
Figure 2 (A) PLA-honeycomb-patterned surface. Image size is 5 μm × 5 μm, scale bar 1 μm, and height bar is 100 nm. (B) E.coli on PLA-honeycomb patterned surface. Image size 15 μm × 15 μm, scale bar is 3 μm, and height bar is 800 nm, (C) Phase image of E.coli on PLA-honeycomb patterned (same surface as image B). Image size is 5 μm × 5 μm. (D) 3D representation of the two E.coli cells from image B (magnification). All images were measured in imaging buffer. Bacterial adhesion time was 30 minutes.
E.coli is one of the best characterized microorganisms as of today and was thus used as model bacteria in the following experiments. Investigating the adhesion of E.coli cells to the substrates; images of E.coli attached to a PLA-honeycomb-patterned surface were recorded in 10 mM HEPES buffer at pH 7.0 (imaging buffer), see Figure 2. Figure 2A shows the PLA-honeycomb-polymer surface before the addition of E.coli cells. The honeycomb-patterned surface shows a very narrow pore size distribution (see also profiles). In Figure 2B seven to eight E.coli cells are distributed on a surface area of 15 μm × 15 μm. The width of the cells is approx. 1 μm, the length is approx. 2.5 μm, and the height is 0.5 μm (see profiles Figure 2B). The feature located next to the scale bar is probably made up of two cells as it is twice the length of the E.coli cell above it and shows a faint separation line in the middle. A more detailed image of one E.coli cell is presented in Figure 2C. The image is a phase image, which enhances the details of the bacterial surface. The surface of the bacteria is very smooth and without any defects, such as grooves or indentations indicating that the cell is still alive. Figure 2D is a three-dimensional representation of a magnification on two E.coli cells in Figure 2B, which further illustrates the smoothness of the bacterial surface.
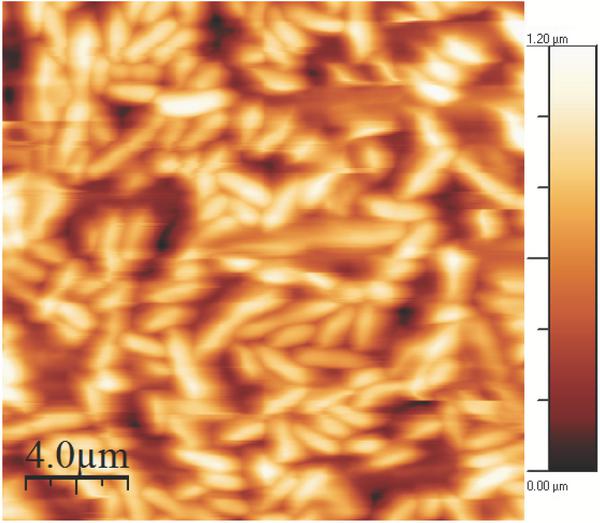
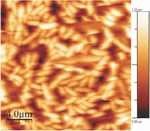
Figure 3 E.coli on a gelatin coated mica surface imaged in imaging buffer.Bacterial adhesion time was 5 minutes. The image is 20 μm × 20 μm, the height bar is 1.2 μm.
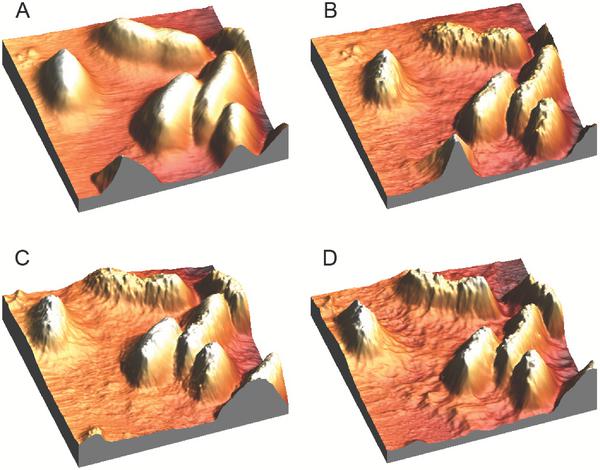
Figure 4 3D AFM height images of E.coli on a gelatin coated surface imaged in imaging buffer. (A) is before the addition of indolicidin, (B) is 20 minutes after the addition of indolicidin, and image (C) and (D) are after approx. 100 and 220 minutes of continuous scanning on the same surface as image A, respectively. Bacterial adhesion time was 1 minute. All images are 8 μm × 8 μm; the height scale is 1.2 μm.
The gelatin coated mica surfaces were tested for E.coli attachment. Images were recorded in imaging buffer, see Figure 3. After 5 minutes of adhesion on a gelatin coated surface, E.coli cells were tightly packed and distributed over the entire measured surface comprising 20 μm × 20 μm. All cells were rod shaped and varied slightly in length, ranging from approx. 2 μm to 3 μm. The density of the cells on the gelatin surface was however too high to study the indolicidin-induced action on the membrane surface. A lower surface density of E.coli cells was achieved using an adhesion time of one minute and images were recorded in imaging buffer, see Figure 4.
Figure 5 3D AFM phase images of E.coli on a gelatin coated surface imaged in imaging buffer (same surfaces as in Figure 4). (A) is before the addition of indolicidin, (B) is 20 minutes after the addition of indolicidin, and image (C) and (D) are after approx. 100 and 220 minutes of continuous scanning on the same surface as image A, respectively. Bacterial adhesion time was 1 minutes. All images are 8 μm × 8 μm, height is 1.2 μm.
In Figure 4A, four whole E.coli cells are observed as well as 3 minor parts of cells. The cells are all rod shaped with lengths around 2.5-3.0 μm as observed in the other AFM scans. Their heights ranges from 0.6-0.8 μm. Figure 4A shows E.coli cells prior to the addition of indolicidin and is obtained after approx. 1 hour of continuous scanning on the same area to obtain a stable image before injecting indolicidin. Furthermore, stability images of E.coli cells in buffer have also been obtained by scanning continuously for 2 hours (data not shown). After the addition of indolicidin, the topography of the cells and the substrate changed notably. The same surface was scanned continuously for approx. 220 minutes in the presence of indolicidin, however the changes in topography are most pronounced after the injection of indolicidin. It should be noted that a second injection of indolicidin was performed after approx. 120 minutes of continuous scanning. Importantly, it was observed that indolicidin very clearly changed the bacterial surface; after addition of indolicidin the roughness of the topography increased. Furthermore, after the addition of indolicidin, some sort of debris is observed on the substrate surface in between the cells and the amount of debris is increasing with time. Before the addition of indolicidin, the surface heights and roughness of the substrate is around 20–30 nm and after 220 minutes in the presence of indolicidin, it has increased to approx. 80–100 nm (data not shown). It is noteworthy that the amount of debris is largest in the surrounding of the cells (compare Figures 4B-D with Figure 4A) – the cell membranes appear to have ‘melted’.
3D phase images of the corresponding height images from Figure 4 are shown in Figure 5. As seen in Figure 5, 3D phase images of the surfaces from Figure 4 provided further details. The effect of injecting indolicidin can be clearly observed when comparing the phase images in Figure 5; indolicidin instantly and dramatically changed the surface of the E.coli cells. Some sort of cellular debris was also present in these images as seen in the height images in Figure 4. Comparing Figures 5A-D, the profiles of the bacterial cells decreases over time; this could be due to a phase change of the membranes.
4 Discussion
E.coli was attached successfully to a honeycomb-patterned surface.The dimensions of the E.coli cells correspond to dimensions stated by others. However, the height of the E.coli cells was lower than the reported heights of immobilized E.coli cells of approx. 600–800 μm [6]. This is due to a strong attraction to the surface which causes a flattening of the bacterial cells. This is an unfavourable effect of the interactions between the honeycomb-patterned surface and the E.coli cells. When forcing the cells to the surface the fluidity of the cell membranes can change and this will not allow for a representative investigation of the interactions between indolicidin and the bacterial cell membranes.
For comparison E.coli was successfully adhered to gelatin coated mica surfaces. Several bacterial species, including E.coli, bind collagen and gelatin (denatured collagen) via specific binding sites on the intact bacterial surface. It is likely that these binding sites, along with electrostatic and hydrophobic interactions, contribute to retaining bacteria on gelatin-coated substrates. Furthermore, adhesion proteins associated with the outer surface and pili of E.coli are likely contributors to the immobilization of the intact bacteria on gelatin-coated mica [4]. The gelatin coated mica surfaces were used in the following experiments with indolicidin and E.coli cells, as these substrates provided higher packing density of E.coli cells compared to PLA-honeycomb-patterned surfaces. This allowed for the observation and quantification of the effect of the peptide acting on several bacterial cells at once.
The cells adhered to the gelatin surface were rod shaped with dimensions that matched the sizes found in literature for E.coli cells [5, 6]. The height profiles from Figure 3 show that the longer features are probably two E.coli cells oriented end to end. This could be dividing cells as the cells have been harvested in the logarithmic phase prior to the immobilization. The bacterial cells imaged in liquid media are rod shaped and smooth compared to results obtained by others measuring E.coli cells in air. In air, highly detailed images visualizing surface topography and roughness are often presented. Additionally, bacterial flagella are seen when imaged in air. These structures are especially evident in amplitude images. The absence of flagella in the images obtained in liquid indicates that the flagella may not be rigidly immobilized or forces exerted by the scanning probe tip may be repositioning this structure [40]. The decreased membrane surface roughness observed in liquid media could be due to membrane fluidity [41]. In contrast, images in air are dramatically different, bacterial cells imaged in air have a flattened scalloped shape with raised edges. These features are likely due to dehydration effects, which can be overcome imaging in liquid.
From Figure 3 it was observed that 5 minutes of adhesion time resulted in a high packing density of E.coli cells on the substrate surface. Under these immobilization conditions, multiple layers of bacteria were observed making AFM measurements difficult to interpret with regards to visualization of peptide-bacterial interactions. This problem was overcome by decreasing the adhesion time and diluting the bacterial concentration as shown in Figure 4. Even after in situ AFM imaging the bacterial cells on the gelatin surface for a couple of hours, the cells remained attached to the support and the shape, size, and height of the cells remained unaffected (data not shown). After the injection of indolicidin, the topography of the cells and the substrate changed notably as can be seen in Figure 4. The roughness of the topography increased as a function of time and some sort of debris was observed on the substrate surface; this could be due to leakage of E.coli cytosol, loss of lipids from the membrane, or peptide molecules carpeting the surface. Combining these observations with AFM studies performed on model membrane systems composed of different phospholipid bilayers indicate that the bacterial cell membranes have become more fluid or ‘melted’ [38].
As seen in Figure 5 the corresponding 3D phase images of the surfaces from Figure 4 provided further details: the effect of injecting indolicidin was further seen in the phase images in Figure 5; indolicidin instantly and dramatically changed the surface behaviour of the E.coli cells. The debris observed in the height images was also observed in the phase images. Before the addition of indolicidin, the bacterial cell membrane is harder compared to after injection of indolicidin, which is seen by the decrease in height appearance of the bacterial cells. This is explained by a decreased suppression of the cantilever amplitude after addition of indolicidin due to a higher degree of fluidity of the cell membrane. All these observations further indicate a fluidization of the membrane. Experiments performed with E.coli are comparable to results obtained with model membranes composed of phospholipids, where fluidization is also believed to occur and therefore it is believed that model membranes are suitable representatives for real membrane systems [38].
5 Conclusion
A comparison of gelatin coated surfaces and PLA-honeycomb patterned mica surfaces was performed with a view to attach E.coli cells for further AFM studies. One main difference between the gelatin coated mica surfaces and the PLA-honeycomb patterned matrices is the softness of the surface. Gelatin is a soft material and will therefore provide a softer surface in which the E.coli cells can slightly submerge leading to a better adhesion, whereas PLA constitute a harder surface implying a decreased bacterial cell density. From a statistical point of view, gelatin coated surfaces are preferred as these make it possible to visualize indolicidin acting on several cells at once and compare these interactions between single cells. PLA-honeycomb patterned surfaces allow for monitoring of isolated single cells. However, both surfaces were suitable for continuous AFM imaging of bacterial cells in liquid.
Mica surfaces coated with either gelatin or a PLA-honeycomb-patterned polymer surface have shown sufficient surface roughness suitable for adhesion of E.coli cells for continuous AFM imaging in liquid. Images obtained using in situ AFM operating in liquid media showed that the E.coli cells had a rod shape and smooth surface. After injection of indolicidin, the bacterial membranes were observed to be distorted and seemed to have ‘melted’, which proceeded as a function of time, indicating fluidization of the cell membrane. This mechanism of action of indolicidin is supported by previous studies using different phospholipid model membranes [38]. Only E.coli cells were attached to solid substrates and visualized using in situ AFM.
It is observed, that indolicidin disrupts the cellular membranes and changes the fluidity notably. However, the E.coli cells were not ruptured as could be expected due to pore formation and disruption of the osmotic pressure. The shape of the cells remains relatively intact upon the addiotion of indolicidin, which could indicate a possible intracellular target killing mechanism of indolicidin interacting with E.coli cells.
Acknowledgments
This work was generously supported by grants from the Spar Nord Foundation and Obels Family Fund.
References
[1] Dufrêne, Y. F. (2004). Refining our perception of bacterial surfaces with the atomic force microscope. Journal of bacteriology, 186(11),
[2] Dufrene, Y. F. (2001). Application of atomic force microscopy to microbial surfaces: from reconstituted cell surface layers to living cells. Micron, 32(2), 153–165.
[3] Radmacher, M., Tillamnn, R. W., Fritz, M. and Gaub, H. E. (1992). From molecules to cells: imaging soft samples with the atomic force microscope. Science, 257(5078), 1900–1905.
[4] Sullivan, C. J., Morrell, J. L., Allison, D. P. and Doktycz, M. J. (2005). Mounting of Escherichia coli spheroplasts for AFM imaging. Ultramicroscopy, 105(1–4), 96–102.
[5] Doktycz, M. J., Sullivan, C. J., Hoyt, P. R., Pelletier, D. A., Wu, S. and Allison, D. P. (2003). AFM imaging of bacteria in liquid media immobilized on gelatin coated mica surfaces. Ultramicroscopy, 97(1–4), 209–216.
[6] Beckmann, M. A., Venkataraman, S., Doktycz, M. J., Nataro, J. P., Sullivan, C. J., Morrell-Falvey, J. L. and Allison, D. P. (2006). Measuring cell surface elasticity on enteroaggregative Escherichia coli wild type and dispersin mutant by AFM. Ultramicroscopy, 106(8–9), 695–702.
[7] Boonaert, C. J. P., Rouxhet, P. G. and Dufrêne, Y. F. (2000). Surface properties of microbial cells probed at the nanometre scale with atomic force microscopy. Surface and Interface Analysis: An International Journal devoted to the development and application of techniques for the analysis of surfaces, interfaces and thin films, 30(1), 32–35.
[8] Micic, M., Hu, D., Suh, Y. D., Newton, G., Romine, M. and Lu, H. P. (2004). Correlated atomic force microscopy and fluorescence lifetime imaging of live bacterial cells. Colloids and Surfaces B: Biointerfaces, 34(4), 205–212.
[9] Ge, G., Han, D., Lin, D., Chu, W., Sun, Y., Jiang, L. and Wang, C. (2007). MAC mode atomic force microscopy studies of living samples, ranging from cells to fresh tissue. Ultramicroscopy, 107(4–5), 299–307.
[10] Ostendorf, F., Schmitz, C., Hirth, S., Kühnle, A., Kolodziej, J. J. and Reichling, M. (2008). How flat is an air-cleaved mica surface?. Nanotechnology, 19(30), 305705.
[11] Missirlis, Y. F. and Katsikogianni, M. (2007). Theoretical and experimental approaches of bacteria-biomaterial interactions. Materialwissenschaft und Werkstofftechnik: Entwicklung, Fertigung, Prüfung, Eigenschaften und Anwendungen technischer Werkstoffe, 38(12), 983–994.
[12] Grantham, M. C., Dove, P. M. and Dichristina, T. J. (1997). Microbially catalyzed dissolution of iron and aluminum oxyhydroxide mineral surface coatings. Geochimica et Cosmochimica Acta, 61(21), 4467–4477.
[13] Scheuerman, T. R., Camper, A. K. and Hamilton, M. A. (1998). Effects of substratum topography on bacterial adhesion. Journal of colloid and interface science, 208(1), 23–33.
[14] Edwards, K. J. and Rutenberg, A. D. (2001). Microbial response to surface microtopography: the role of metabolism in localized mineral dissolution. Chemical Geology, 180(1–4), 19–32.
[15] Katsikogianni, M. and Missirlis, Y. F. (2004). Concise review of mechanisms of bacterial adhesion to biomaterials and of techniques used in estimating bacteria-material interactions. Eur Cell Mater, 8(3), 37–57.
[16] Yao, X., Walter, J., Burke, S., Stewart, S., Jericho, M. H., Pink, D. and Beveridge, T. J. (2002). Atomic force microscopy and theoretical considerations of surface properties and turgor pressures of bacteria. Colloids and Surfaces B: Biointerfaces, 23(2–3), 213–230.
[17] Fukuhira, Y., Kitazono, E., Hayashi, T., Kaneko, H., Tanaka, M., Shimomura, M. and Sumi, Y. (2006). Biodegradable honeycomb-patterned film composed of poly (lactic acid) and dioleoylphosphatidylethanolamine. Biomaterials, 27(9), 1797–1802.
[18] Y. Fukuhira, H. Kaneko, M. Tanaka, M. Yamaga, S. Yamamoto, M. Shimonura, Colloids and Surfaces A: Physicochem. Eng. Aspects 313–314 (2008) 220.
[19] Toke, O. (2005). Antimicrobial peptides: new candidates in the fight against bacterial infections. Peptide Science: Original Research on Biomolecules, 80(6), 717–735.
[20] Boman, H. G. (2003). Antibacterial peptides: basic facts and emerging concepts. Journal of internal medicine, 254(3), 197–215.
[21] Hancock, R. E. and Scott, M. G. (2000). The role of antimicrobial peptides in animal defenses. Proceedings of the national Academy of Sciences, 97(16), 8856–8861.
[22] Bradshaw, J. P. (2003). Cationic antimicrobial peptides. BioDrugs, 17(4), 233–240.
[23] Makovitzki, A., Avrahami, D. and Shai, Y. (2006). Ultrashort antibacterial and antifungal lipopeptides. Proceedings of the National Academy of Sciences, 103(43), 15997–16002.
[24] Shai, Y. (2002). Mode of action of membrane active antimicrobial peptides. Peptide Science: Original Research on Biomolecules, 66(4), 236–248.
[25] Sawyer, J. G., Martin, N. L. and Hancock, R. E. (1988). Interaction of macrophage cationic proteins with the outer membrane of Pseudomonas aeruginosa. Infection and immunity, 56(3), 693–698.
[26] Subbalakshmi, C., Krishnakumari, V., Sitaram, N. and Nagaraj, R. (1998). Interaction of indolicidin, a 13-residue peptide rich in tryptophan and proline and its analogues with model membranes. Journal of biosciences, 23(1), 9–13.
[27] Matanic, V. C. A. and Castilla, V. (2004). Antiviral activity of antimicrobial cationic peptides against Junin virus and herpes simplex virus. International journal of antimicrobial agents, 23(4), 382–389.
[28] Bera, A., Singh, S., Nagaraj, R. and Vaidya, T. (2003). Induction of autophagic cell death in Leishmania donovani by antimicrobial peptides. Molecular and biochemical parasitology, 127(1), 23–35.
[29] Bhargava, A., Osusky, M., Hancock, R. E., Forward, B. S., Kay, W. W. and Misra, S. (2007). Antiviral indolicidin variant peptides: Evaluation for broad-spectrum disease resistance in transgenic Nicotiana tabacum. Plant science, 172(3), 515–523.
[30] Shaw, J. E., Alattia, J. R., Verity, J. E., Privé, G. G. and Yip, C. M. (2006). Mechanisms of antimicrobial peptide action: studies of indolicidin assembly at model membrane interfaces by in situ atomic force microscopy. Journal of structural biology, 154(1), 42–58.
[31] Ramamoorthy, A., Thennarasu, S., Lee, D. K., Tan, A. and Maloy, L. (2006). Solid-state NMR investigation of the membrane-disrupting mechanism of antimicrobial peptides MSI-78 and MSI-594 derived from magainin 2 and melittin. Biophysical journal, 91(1), 206–216.
[32] Hsu, C. H., Chen, C., Jou, M. L., Lee, A. Y. L., Lin, Y. C., Yu, Y. P. and Wu, S. H. (2005). Structural and DNA-binding studies on the bovine antimicrobial peptide, indolicidin: evidence for multiple conformations involved in binding to membranes and DNA. Nucleic acids research, 33(13), 4053–4064.
[33] Zasloff, M. (2002). Antimicrobial peptides of multicellular organisms. nature, 415(6870), 389.
[34] Chan, D. I., Prenner, E. J. and Vogel, H. J. (2006). Tryptophan-and arginine-rich antimicrobial peptides: structures and mechanisms of action. Biochimica et Biophysica Acta (BBA)-Biomembranes, 1758(9), 1184–1202.
[35] Yau, W. M., Wimley, W. C., Gawrisch, K. and White, S. H. (1998). The preference of tryptophan for membrane interfaces. Biochemistry, 37(42), 14713–14718.
[36] Dougherty, D. A. (1996). Cation-p interactions in chemistry and biology: a new view of benzene, Phe, Tyr, and Trp. Science, 271(5246), 163–168.
[37] Schibli, D. J., Epand, R. F., Vogel, H. J. and Epand, R. M. (2002). Tryptophan-rich antimicrobial peptides: comparative properties and membrane interactions. Biochemistry and cell biology, 80(5), 667–677.
[38] Askou, H. J., Jakobsen, R. N. and Fojan, P. (2008). An atomic force microscopy study of the interactions between indolicidin and supported planar bilayers. Journal of nanoscience and nanotechnology, 8(9),
[39] Horcas, I., Fernández, R., Gomez-Rodriguez, J. M., Colchero, J. W. S. X., Gómez-Herrero, J. W. S. X. M. and Baro, A. M. (2007). WSXM: a software for scanning probe microscopy and a tool for nanotechnology. Review of scientific instruments, 78(1), 013705.
[40] Bolshakova, A. V., Kiselyova, O. I., Filonov, A. S., Frolova, O. Y., Lyubchenko, Y. L. and Yaminsky, I. V. (2001). Comparative studies of bacteria with an atomic force microscopy operating in different modes. Ultramicroscopy, 86(1–2), 121–128.
[41] Robichon, D., Girard, J. C., Cenatiempo, Y. and Cavellier, J. F. (1999). Atomic force microscopy imaging of dried or living bacteria. Comptes Rendus de l’Academie des Sciences-Series III-Sciences de la Vie, 322(8), 687–693.
Biographies

Peter Fojan received his Ph.D. in Biotechnology at the University of technology Graz, Austria in 1997. He initially worked on industrial genetics of eukaryotic organisms. During his postdoc time at Aalborg University at the Department of Biotechnology he moved into the area of protein physics and molecular modelling. With the startup of Nanotechnology at AAU he moved to the Department of Physics and Nanotechnology where he became an Associate Professor in 2009. His research interests are centered around biological and small molecules and their interactions with cells and surfaces in general, for medical, sensor applications and as antibacterial agents.
Journal of Self-Assembly and Molecular Electronics, Vol. 6_1, 13–34.
doi: 10.13052/jsame2245-4551.6.002
This is an Open Access publication. © 2018 the Author(s). All rights reserved.