Salt Bridges Regulate in Silico Dimers Formation for β2-Microglobulin Amyloidogenic Variants
Maria Celeste Maschio1,2, Giorgia Brancolini2,*, and Stefano Corni2,3
1Department of Physics, University of Modena and Reggio Emilia, Via Campi 213/a, 41125 Modena, Italy
2Center S3, CNR-NANO Institute of Nanoscience, Via Campi 213/A, 41125 Modena, Italy
3Department of Chemical Science, University of Padova, Via Marzolo 1, 35131 Padova, Italy
E-mail: giorgia.brancolini@nano.cnr.it
∗Corresponding Author
Received 05 November 2018; Accepted 21 December 2018;
Publication 02 January 2019
Abstract
β2-microglobulin is a paradigmatic amyloidogenic protein responsible for dialysis-related amyloidosis, a disease associated to long-term hemodialyzed patients and characterized by accumulation of amyloid deposits in the osteoarticular tissues. In the early stages of amyloid fibril formation, β2-microglobulin associates into dimers and higher oligomers, but clarifications are still needed for the triggering conditions, mechanisms and specificity of dimer formation. To characterize the dimeric association process, the protein-protein interactions between three different species are investigated: namely, the native protein and the two amyloidogenic variants ΔN6 and D76N. The dimerization process is rationalized relying on state of the art computational methods. A comparative mechanism for how different mutations in the three variants can affect protein dimerization and thus fibril formation is proposed. The number of salt bridges involved at the protein-protein interface correlates with the degree of amyloidogenicity of each individual species. The findings can offer possible strategies in controlling the dimerization mechanism based on different β2-microglobulin protein mutations, which have significant roles in the fibrillogenical process.
Keywords
- amyloidosis
- molecular dynamics
- protein aggregation
- β2-microglobulin
1 Introduction
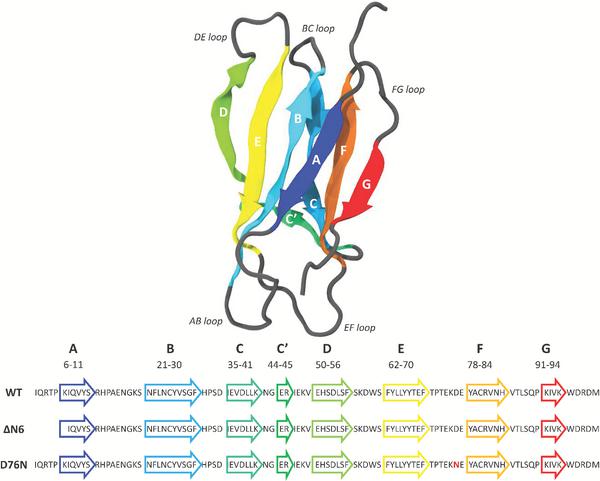
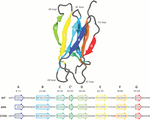
β2-microglobulin (β2-m) pertains to the immunoglobulin family and it is the light chain of the major histocompatibility complex (MHC) class I [1]. It is a 99 residue protein, characterized by seven anti-parallel β strands folded in a β sandwich, stabilized by a disulfide bridge between Cys25 and Cys80 (Figure 1). When released in monomeric form, β2-m loses the stability given by the interaction with MHC class I [2] and it can lead to Dialysis Related Amyloidosis (DRA), a systemic amyloidosis affecting patients undergoing long-term hemodialysis. In fact, biochemical studies [3–6] have reported that β2-m is the constituent of amyloid deposits of patients treated with chronic hemodialysis. The wild type β2-m requires particular conditions in order to form amyloid fibrils in vitro, as the presence of ex vivo amyloid seeds [7] or Cu2+ ions [8]. Further studies on different types of tissues have shown that ΔN6, the truncated species lacking the six N-terminal (N-term) residues, is also significantly present [9–11] (around 30% of ex vivo amyloid deposits) in patients affected by DRA (Figure 1, where one-letter code sequence highlight the N-term cleavage). There are different and controversial findings regarding the mechanisms through which ΔN6 can prime the fibrillogenical behavior of the wild type [12, 16], but it is widely recognized that the truncated variant alone is able in nucleating fibrillogenesis in physiological conditions. While no other major post-translational modifications are apparently present in natural fibrillar β2-m, few years ago the first natural amyloidogenic variant of β2-m was discovered in a French family where all the heterozygous carriers of the mutation presented a multi-visceral amyloid deposit [13, 14]. The replacement of aspartic acid in position 76 with the asparagine (D76N) leads to a highly amyloidogenic protein, able to fibrillate under physiological condition [15] (Figure 1, where one-letter code sequence highlights the mutation position in red).
Figure 1 Backbone structure of the native β2-microglobulin (PDB entry: 1JNJ) as obtained from NMR determination [31] in aqueous solution.
Amyloid formation of β2-m under physiological pH conditions (around pH 7.0) has been shown to start from the fully folded native protein state [17]. β2-m fibrillation can be triggered by conformational rearrangement of the natively folded structure [18, 19], but whether the fibrillogenical mechanism requires rearrangement to a native-like species or more highly unfolded species, or both, under different solution conditions remained unclear [4, 19–21]. Furthermore, it is not known whether the transition to the amyloidogenic intermediate may be favored by native protein dimerization or not. In the present work we rely on native conformation as a starting point to perform an enhanced conformational search of dimeric protein adducts.
Atomistic molecular dynamics is a powerful tool that can effectively complement experimental studies on this topic [23, 24]. The ability of β2-m in forming dimers under native conditions and the subsequent aggregation into tetramers has been modeled in different previous works [19, 25, 26]. In addition, dimeric units have been suggested as fundamental blocks in the construction of the mature fibrils [27]. In this work, we perform a comparative analysis of the interactions between dimers of β2-m wild type (WT) and of amyloidogenic variants ΔN6 and D76N, starting from native conformations representative of the protein in physiological conditions. All calculations involve simulations at multiple levels spanning from docking [29] to conventional atomistic molecular dynamics (MD) and Temperature Replica Exchange MD (T-REMD) [30]. The results show that the dimer interface of the WT β2-m is more unsteady than ΔN6 and D76N variant interfaces, in agreement with the enhanced amyloidogenicity of such mutants. A mechanism of sequential formation (“zipper”-like) of the salt bridges at the protein-protein interface is proposed, based on the current in silico results. The role of salt bridges in stabilizing the “zipped” dimer for each mutated species is addressed by tracking their formation during classical MD and then assessing their stability also by means of T-REMD simulations. The persistence in time of salt bridges involved at the protein-protein interface correlates with the different degree of amyloidogenicity between the compared species.
2 Results and Discussions
2.1 Exploring Protein-protein Binding Interfaces
The dimeric association of wild-type β2-m or its variants in solution has been frequently observed [10, 22, 32, 33]. In order to provide a complementary approach to the interpretation of available experimental data, a preliminary docking to build protein-protein dimers is performed. We simulate dimers of WT, ΔN6 and D76N. The prediction of protein-protein encounter complexes is performed by applying the rigid-body docking as coded in SDA7 [29]. These structures are generated by running around 5000 Brownian Dynamics (BD) simulations during which the solutes are modeled as rigid bodies in presence of implicit solvent. The adsorption free energies of the protein-protein encounter complexes obtained during the BD simulation are computed and the trajectories are clustered with the single linkage method to identify genuinely different protein orientations [34]. Flexible refinement of selected representative structures is hence done by molecular dynamics (MD) simulations in explicit solvent. The advantage of using BD is that it mimics efficiently the physical process of diffusional association of the unbound proteins whereas the atomistic refinement is accounting for the protein conformational flexibility in MD upon association.
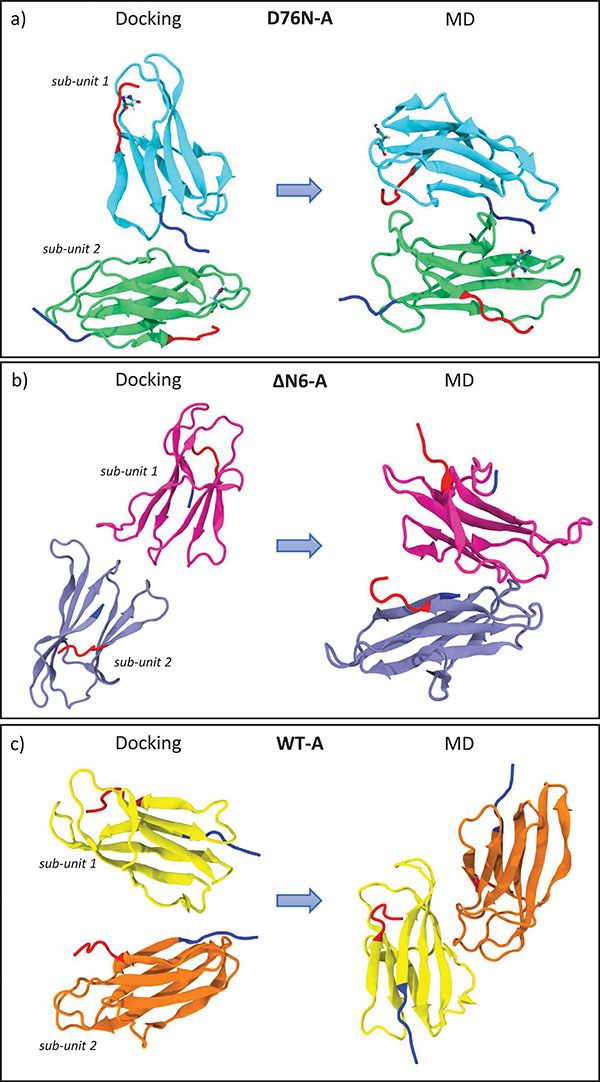
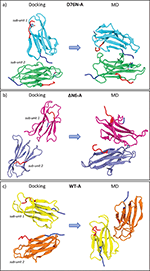
The same procedure is repeated for each variant: D76N variant yields one very populated complex which is representative for almost 97% of the total encounter complexes, whereas ΔN6 and WT dimers yield two different reciprocal orientations and four different orientations, respectively. Statistical data on the resultant docked complexes are reported in SI, in Table ST1. For ΔN6 we select the most populated complex (ΔN6-A) representing the 70% of the total complexes, whereas for WT the first encounter complex (WT-A) is representative of the 62%. During docking, the interaction energy between the two proteins is described by the electrostatic interaction term, UEP, dominated by the contribution of the charged side chains, and the non-polar hydrophobic interactions of the protein, . Binding between all dimeric complexes is driven mostly by non-polar hydrophobic energy terms. D76N-A dimer shows the most favorable total binding energy between all dimers. The difference between the binding energies in complexes of D76N-A and ΔN6-A is of ∼13 kT whereas between complexes of D76N-A and WT-A is of ∼9 kT, respectively. In the case of D76N-A dimer, different contact residues of each monomer forming the dimer are involved at the protein-protein binding interface: sub-unit 1 involves N-term (THR4, PRO5), BC loop (HIS31) and FG loop (THR86, SER88) and sub-unit 2 involves B strand (PHE22), CD loop (ILE46, GLU47, LYS48), D strand (GLU50, ASP53) and E strand (TYR67, GLU69) (see Figure 1 to identify strands and loops). In the case of ΔN6-A dimer the electrostatic interaction is way weaker and the dimeric interface is mainly stabilized by non-polar hydrophobic interactions. Residues involved within the binding interface for sub-unit 1 are in BC loop (SER33), DE loop (LEU54, TRP60), D strand (PHE62) and for sub-unit 2 are in BC loop (HIS31, PRO32, ASP34) and FG loop (THR86). Concerning the WT-A protein at physiological pH, the dimeric interface is stabilized mainly by non-polar hydrophobic interactions but also the electrostatic energy term partially contributed to binding. Binding patches are involving residues in N-term (LYS3), DE loop (SER61) and G strand (LYS91) for sub-unit 1 and D strand (ASP53, SER55), DE loop (PHE56, LYS58) for sub-unit 2. In the case of the WT-A, protein docking results are in agreement with NMR experimental data providing evidences of the existence of dimeric adducts in pure water with the N-term segment and DE loop involved at the dimeric interface[22]. The DE loop and the contiguous D and E strands are involved in interfaces for all the investigated species, confirming the relevance of this portions in interactions at the beginning of the aggregation [24, 28, 35].
Starting with the most representatives dimeric complexes obtained from rigid-body BD docking, the stability of each interface is examined by using four MD runs (different by initial atomic velocities) of 400 ns each in explicit water. Given the size of the protein surface, a large number of different interactions is expected during MD, as depicted in Figure S1 in SI and here summarized in Table 1. The four independent MD runs performed on mutated dimers D76N and ΔN6 generate ensembles of conformations characterized by more extended binding interfaces with respect to the initial ones. The initial D76N-A dimer resulting from docking is able to relax into novel interface in two cases over four. Instead, ΔN6-A dimer conserves its binding patch in three cases over four. Partially this is due to the fact that population and stability of dimeric interfaces can be affected differently by relaxation during MD refinement and can also depend on mutated regions i.e. EF-loop for D76N and N-term deletion for ΔN6. In WT the binding interface is involving a smaller number of residues compared to the other mutated species.
Relaxed conformations emerging from MD show distinctive features as the formation of salt bridges in interaction interface for both D76N and ΔN6. In our findings, the extended interface between sub-units are named as “zipped” dimers, where with “zipping” mechanism we are referring to the pivotal role of a number of charged residues forming stable “salt bridge zipper” between the two monomers. During MD, a spatially sequential formation of such bridges is found. Asparagine substitution with aspartate at residue 76 increases the tendency in forming novel salt bridges within the two sub-units of D76N-A. The same capability of forming “zipped” interfaces is observed in ΔN6-A. On the contrary, formation of salt-bridges is not observed for WT-A. This is in line with the experimental evidence reporting the native protein not be able to form amyloid fibrils in vitro in physiological conditions (i.e. protein charge state at neutral pH) [12, 19, 36]. Given the different behavior of D76N and ΔN6 variants with respect to WT, we first focus on the “zipped” dimers in order to rationalize their mechanism and potential role in the formation of stable dimers, which could be relevant for early stages in fibrillation. A last section is describing briefly the “non-zipped” WT. Considering Table 1 and Figure S1 in SI, the analyzed dimers are a1 for D76N, b1 for ΔN6 and c1 for WT.
Table 1 Orientations of D76N-A, ΔN6-A and WT-A dimers obtained following the refinement MD runs. Four indipendent simulations (long 400 ns at 300 K) are performed starting from the most representative protein-protein complexes. The final global orientation of the systems after MD simulation is reported, together with number of formed salt bridges in the new interaction interfaces
| Dimeric Complex | Number of SBs | Global Orientation | Fig. S1 |
| D76N Dimer 1 | 3 | converted to zipped | (a1) |
| D76N Dimer 2 | 1 | converted to zipped | (a2) |
| D76N Dimer 3 | 0 | stable | (a3) |
| D76N Dimer 4 | 1 | stable | (a4) |
| ΔN6 Dimer 1 | 4 | converted to zipped | (b1) |
| ΔN6 Dimer 2 | 1 | stable | (b2) |
| ΔN6 Dimer 3 | 0 | stable | (b3) |
| ΔN6 Dimer 4 | 0 | stable | (b4) |
| Wild Type Dimer 1 | 1 | rearranged | (c1) |
| Wild Type Dimer 2 | 1 | rearranged | (c2) |
| Wild Type Dimer 3 | 0 | stable | (c3) |
| Wild Type Dimer 4 | 0 | stable | (c4) |
Table 2 Time formation of salt bridges during simulation
| Dimeric Complex | Salt Bridge Residues | Time (ns) | |
| sub-unit 1 | sub-unit 2 | ||
| D76N | Asp34 | Arg97 | 84 |
| Arg3 | Glu69 | 86 | |
| His84 | Asp98 | 157 | |
| ΔN6 | Asp34 | Lys91 | 61 |
| His51 | Asp59 | 62 | |
| Glu36 | Ile7 | 142 | |
| Glu44 | Arg12 | 144 | |
| Wild Type | Lys75 | Glu50 | 165 |
2.2 Dimerization Mechanism
The dimerization mechanism is discussed and analyzed for the three species of β2-m, separately. The stability of our simulations is assessed by the Root Mean Square Deviation (RMSD) calculated for the protein backbone without the terminal tails and by the radius of gyration Rg (see SI, Figure S2).
2.2.1 D76N “zipped” dimer
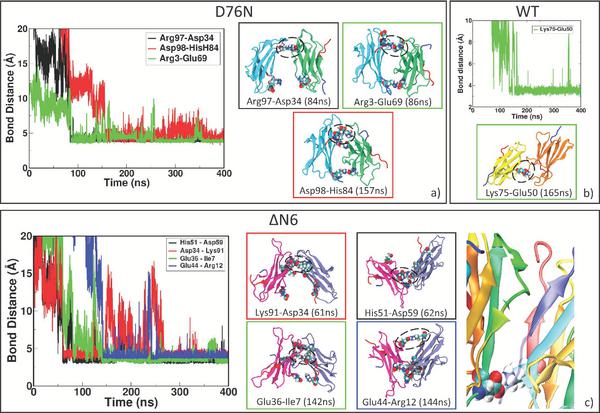
In D76N dimer, the two monomers rearrange after sliding during the formation of three salt bridges (Table 2), ending in a larger number of intermolecular contacts at the interface (Figure 2a). The “salt bridge zipper” between the two monomers starts with a bond involving ASP34-ARG97 at time 84 ns followed by the formation of ARG3-GLU69 couple at 86 ns and the final slippage is assessed at 157 ns with a salt bridge between HIS84-ASP98 (Figure 3a).
At the end of simulation, the protein-protein binding interface involves the N and C-term of one monomer and leaves the terminal tails of the second monomer free to bind to another identical protein. As reported in the SI in Table ST2, the final interface relies on residues belonging to the regions identified as crucial for amyloidogenesis: more specifically, sub-unit 1 is involving residues of N-term, AB loop, C-term and of DE loop, very unstable segment of D76N β2-m variant. Also EF loop and F strand, that contains the single point mutation, are interacting with the sub-unit 2 in the final “zipped” dimer. The sub-unit 2 is involving residues belonging to complementary protein region respect to sub-unit 1. Our results are in line with previous modeling work at a more coarse grained level description [37], in which the authors studied dimers of D76N and put emphasis on the important role of DE loop for dimerization at physiological pH conditions, indicating as aggregation hot spots PHE56, TRP60, TYR63, ASP59, PHE62, TYR66 and also predicted as relevant residues LYS94 and TRP95. Based on a series of independent fully relaxed atomistic simulation of D76N dimers, we find a binding interface with strong presence of residues pertaining to DE loop and also residues from the C-term tail as predicted. Such interface is further supported by recent experimental evidence reporting that a specific nanobody (Nb24) is able to prevent amyloidogenesis of D76N β2-m [38]. The effects of Nb24 binding are observed in the apical AB and EF loops and the C-terminal of D76N, directly involved in the protein self-association of the “zipped” dimer. The binding of an anti-aggregation drug or antibody with anti-amyloidogenic power in those sites would be able of blocking the protein from binding to another protein thus leading to an inhibition of the fibrillation activity, and our results confirm these experiments.
Figure 2 Reciprocal orientations before and after 400 ns of MD: on the left, initial complexes from docking; on the right, final orientations after MD. In panel a) D76N sub-unit 1 is in light blue and sub-unit 2 in light green. In panel b) ΔN6 sub-unit 1 is represented in light magenta and sub-unit 2 in gray. In panel c) WT sub-unit 1 is painted in yellow and sub-unit 2 in orange. For all systems, the N-term tail and the C-term tail are depicted in blue and red color, respectively (VMD visualization tool).
Overall, our simulations suggest that changes applied locally to D76 residue are able to highlight cooperative rearrangements on coupled regions of the protein, resembling allosteric events. In particular, when the interaction between E and F strands becomes weaker with respect to the native interaction due to the replacement of a charged residue D76 with a neutral residue N76, the strands A and B are getting more loosely bound to the protein and tend to fluctuate more freely respect to the WT, favoring the formation of a larger number of salt bridges being formed. Experimental evidences have already suggested that by weakening the interaction between D and E strand through the inclusion of mutations at Trp60 it was possible to affect the strength of the interaction between F and G strands (Trp95), inducing allosteric type behavior on the protein [39].
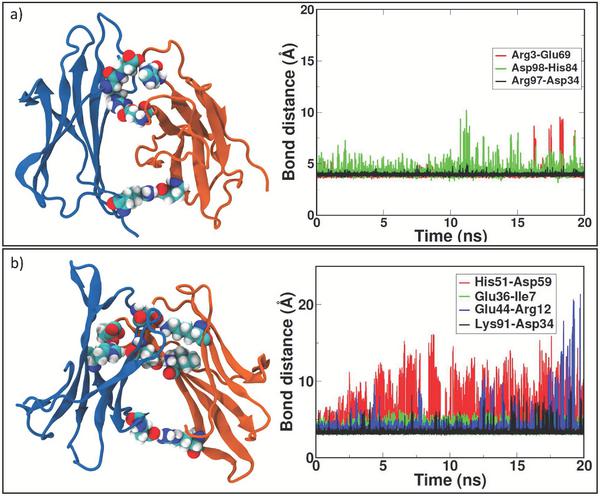
Figure 3 Zipping mechanism and salt bridges formation in dynamics of the three dimers. In each panel, the distances between charged residue and the timing of stable formation are reported. In panel a) formation of the first SB between two identical D76N monomers at 84 ns, the second at 86 ns and the final zipping at 157 ns. In panel b) lone SB formation for WT at 165 ns. In panel c) first SB formation between ΔN6 monomers at 61ns, the second at 62 ns and the final zipping at 144 ns. The last figure in panel c) shows the role of missing N-term tail (transparent blue) in allowing interaction between LYS91 and ASP34.
2.2.2 ΔN6 “zipped” dimer
Once reached stability (SI, Figure S3), it is observed that initial interface involving N-terminal apical region of both sub-units change into a novel “zipped” dimer. During dynamics, larger β-sheet portions of the two sub-units (Figure 2b) play a major role. As reported with more details in SI in Table ST3, involved residues for sub-unit 1 come from BC loop, C’ strand, CD loop, D strand, F strand and from FG loop. For sub-unit 2, involved residues are from A strand, AB loop, B strand, DE loop, E-F strand, FG loop and G strand. In this case, the dimer rearrangement drives tails of sub-unit 2 toward the interface. In a previous work [24], Estacio et al. identified the hotspots for ΔN6 dimer aggregation in PHE30, HIS31 and HIS84. The resulting ΔN6 dimer interface in our simulations includes residues from DE loop and G strand, and also HIS84. Also in the case of ΔN6-A, we observe formation of the “salt bridge zipper” between the two sub-units: in this variant the first salt bridge is formed between residues LYS91 of sub-unit 1 and ASP34 of sub-unit 2. The “zipper”-like mechanism is plotted in Figure 3c. From Table 2, we can see that the formation of this first salt bridge occurs earlier than in any other investigated variants, and in the end it involves a larger number of salt-bridges with respect to the other variants. In fact, the bond between LYS91 of sub-unit 1 and ASP34 of sub-unit 2 occurs at time 61 ns, followed by formation of a second salt-bridge between HIS51 of sub-unit 1 and ASP59 of sub-unit 2 at time 62 ns. After some time to relax the neighboring residues, the closure occurs at time 142 ns and 144 ns by the formation of salt-bridges between residues GLU36 of sub-unit 1 with ILE7 of sub-unit 2 and GLU44 of sub-unit 1 with ARG12 of sub-unit 2. The first electrostatic contact, by which “zipping” mechanism originates, is not observed in any other variant since it is possible thanks to the missing N-term, as depicted in the zoomed structure in Figure 3c. In order to assess the role of ASP34 in the formation of the “zipped” interface, we run a MD simulation starting from dimer ΔN6 in the conformation with the first salt bridge already formed and then mutation D34N is inserted. After 180 ns, the interaction interface is involving BC loop, C strand, CD loop and FG loop for the first sub-unit where there is the mutation. The second sub-unit is involving FG loop, G strand and C terminus, and also few residues from A strand, B strand and DE loop. The simulation shows that, replacing ASP34 with ANS34, complete “zipping” mechanism does not occur (SI, Figure S12).
2.2.3 WT “non-zipped” dimer
Sub-units of WT-A (SI, Figure S4) are able to rearrange but formation of a “salt bridge zipper” is not present as in other variants (Figure 2c). Very likely, the N-tail does not allow any initial contact between charged residues. New dimeric interaction patches (more details in SI, Table ST4) involve AB loop, EF loop, FG loop and most of the strands (A, B, C and F) for sub-unit 1 and AB, CD loops and B, E strands and C-terminal. We observe that, at the beginning of MD, both the monomers involve DE loop in the binding interface, but during dynamics they loose reciprocal interaction occuring via such loop. In fact, this portion of the protein is strongly mobile in WT and binding is observed to be unstable due to the presence of TRP60. Rennella and co-workers [28] indicated BC loop highly reactive in dimerization process for the wild type and then listed several relevant residues: GLN2, ARG3, LYS6 (N-term); SER28, PHE30, SER33, ASP34, GLU36 (BC loop, C strand); LEU54, SER55 (DE loop); VAL85 (FG loop); ASP96 (G strand). Comparing final residues at the interface of WT-A, we find strong participation of N-term, B strand and D strand residues for the initial docking pose, then the interface proposed by other predictions has been lost, maintaining few residues of A strand and C term. For WT, we observe the formation of a lone salt bridge between LYS75 and GLU50 at 165 ns (Figure 3b). Such bond is indeed well conserved during the entire length of the simulation but no additional salt bridges form. The interaction of the involved charged residues [40] highlighted GLU50 as really important residue in D-E strand strand interaction in driving anti-parallel rearrangement, while LYS75 in EF loop is very active in the dimer formation as it is more solvent accessible.
2.3 Hydrophobic Area and Structural Rearrangements
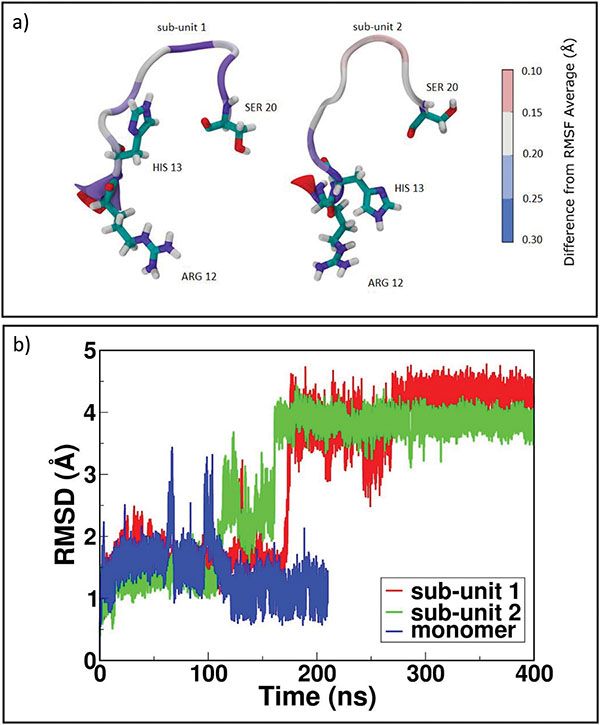
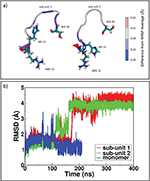
In order to rationalize the dimerization mechanisms, it is crucial to analyze each factor that may confer enhanced amyloidogenic propensity to β2-m variants. Changes in hydrophobicity and in secondary structure of the protein are the most important factors [41]. In this section we discuss how the formation of a dimeric interfaces is accompanied by changes of these two features of protein aggregates. The variation of hydrophobic Solvent Accessible Surface Area (SASA) is studied to investigate the role of hydrophobic forces in driving dimeric rearrangement during relaxation of MD simulations. Analyzing the three dimers (as reported in plot of SASA in SI, Figures S5 and Table ST5), we observe that the formation of D76N zipped dimers is accompanied by a larger variation of hydrophobic SASA, with a decrease of around 6 nm2. On the contrary, ΔN6 and WT dimerization processes are paired with even lower variation of the hydrophobic SASA, showing an almost constant behavior of hydrophobic forces that are not contributing significantly in the overall rearrangement. For the cleaved variant ΔN6, the new configuration tends to favor the interaction between N-term charged residues in both proteins and it is stabilized mainly by favorable electrostatic forces acting in a small N-term region. It results in a variation of 2 nm2 for SASA. A similar behavior is observed for the WT dimer: the sliding between monomers is modest because of the lack of a zipping mechanism hampered by the presence of a flipping tail at N-term. In fact, the last residues of N-term are avoiding the establishment of initial stable electrostatic contacts between the two sub-units. As conformational rearrangements are suspected to be related to the transition towards fibers, we check the structural stability of the portions of each sub-units in every dimers (see SI in Figures S8, S9 and S10). In particular, for β2-m it is conceivable that very limited conformational changes may be sufficient in promoting fibrillogenesis, without strong loop rearrangement and loss of secondary structure [13]. In D76N-A, AB and EF loops present the highest mobility with respect to other portions of the protein but, in general, the RMSD of D76N-A does not deviate from NMR structure 1JNJ. In ΔN6-A, the “zipped” dimer presents a high stability for each portion, although the most interesting rearrangement involves loop AB. Almost contemporarily in both sub-units at time 180 ns (Figure 4b) in sub-unit 1 there is a complete opening toward the solvent, while in sub-unit 2 the new orientation is toward sub-unit 1. AB loop is a quite interesting portion as it is an interaction site between MHC complex and β2-m [42]. It is also relevant that the AB loop of sub-unit 1, which is in direct contact with the surrounding solvent, is experiencing a stronger Root Mean Square Fluctuation (RMSF) of the involved residues with respect to the sub-unit 2 (Figure 4a). A possible explanation for AB loops opening could be found in the closure of the dimer, which through “zipping” is inducing a structural rigidity of those portions of the proteins involved at the interfacial region. To compensate such effect, other portions of the dimer not directly involved in binding might become softer and more mobile, resembling allosteric perturbations observed in the NMR experiments [5].
Figure 4 ΔN6 dimer structural rearrangements. In panel a) RMSF for loop AB in sub-unit 1 and sub-unit 2; in panel b) RMSD for loop AB in ΔN6 dimer sub-units and monomer.
2.4 Stability of Dimeric Interfaces
T-REMD is used to allow a more extensive search of dimeric conformational minima in the Free Energy Surface. We employ a total of 32 replicas, covering a temperature range 290–320 K [48, 50]. The adopted protocol yields an aggregated simulation time of 640 ns. The convergence of the simulation is assessed with stability of protein-protein binding interface which is not changing during the entire length of T-REMD for all performed simulations. A deeper analysis involving existence of salt bridges at the interface can be interpreted as the mechanism by which the interface is stabilized.
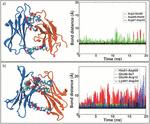
The formed salt bridges are tested by proving stability of the bonding. T-REMD simulations of 20 ns are performed starting from the D76N-A and ΔN6-A “zipped” dimers. Since for WT-A dimer no formation of “salt bridge zipper” is observed, we skip further simulation of this specie. The existence in time of salt bridges at interface is obtained following the replica at the lowest temperature during T-REMD. The interaction patch of dimer D76N-A is shown in Figure 5a: two out of three salt bridges are found to be stable, having a residue-residue distance of 4 Å conserved during the entire simulation (see Table 3). For ΔN6-A, two out of four salt bridges are well conserved during 20 ns of T-REMD. In fact, the distance between charged residues involved in binding interface remains constant between LYS91-ASP34 and GLU36-ILE7, as showed in Figure 5b (black and green lines, respectively). The two remaining salt bridges are experiencing a continuous breaking and reforming, enabling sub-units to slightly rotate around the conserved bonds and change the interaction interface. Nevertheless, conformational rearrangements experienced in the MD are well conserved also in T-REMD simulation, and there are no portions of the protein undergoing relevant structural changes.
Figure 5 First most populated cluster after 20 ns of T-REMD. Residues involved in SB formation are represented with van der Waals radius. In panel a) the formation of three SB within D76N dimers is depicted (left); the SB bond distance is conserved during the entire simulation (right). In panel b) four SB within ΔN6 dimer are highlighted (left); during T-REMD, only two out of four SB are conserved at the end of the simulation (right).
Table 3 Fraction of salt bridges conserved during the T-REMD simulations for the amyloidogenical variants D76N and ΔN6. The threshold distance for the salt bridge is 4 Å and the conservation threshold is above 0.75
| Dimer | Salt Bridge | Fraction of SB Formation |
| D76N | ARG3-GLU69 | 0.89 |
| ASP98-HIS84 | 0.22 | |
| ARG97-ASP34 | 0.76 | |
| ΔN6 | HIS51-ASP59 | 0.70 |
| GLU36-ILE7 | 0.81 | |
| GLU44-ARG12 | 0.71 | |
| LYS91-ASP34 | 0.96 |
3 Conclusions
In this work we propose a comparative mechanism to explain how different mutations of the highly amyloidogenic D76N and ΔN6 β2-m variants can affect dimerization, characterizing intermolecular salt bridges formation. We find in particular that the number of salt bridges involved at the protein-protein interface is directly related to the degree of amyloidogenicity of each individual species (D76N > ΔN6 > WT). Our results show that only D76N and ΔN6 are able to initiate formation of salt bridges between neighboring proteins leading to a “zipping-like” mechanism. On the contrary, for WT formation of a single salt bridge is observed.
Our MD results provide new ideas on the mechanism of formation and stabilization of dimers, suggesting new experiments in the field and contributing in the clarification of how low-weight oligomers are formed in solution and how fibrillogenesis sets up.
4 Methodology
Three proteins are considered: wild-type β2-m, ΔN6 and D76N variants. MD simulations start from the same NMR structure (PDB:1JNJ) [31] upon inclusion of variations: ΔN6 is obtained by cleaving the first six residues and D76N substituting residue ASP76 with ASN. In the case of D76N, the protein is relaxed before docking and a structural sampling in solution is performed for 20 ns of T-REMD, in order to include the possibile conformational changes of the mutated residue at position 76. RMSD comparison with a plethora of other available PDB structures is reported in Table SF1 in SI, with an average RMSD value below 2.8 Å for 1JNJ (PDB entries: 5CS7, 2YXF, 2D4F, 2XKS for WT; 2XKU for ΔN6; 5CSB, 4FXL for D76N). For completeness, a comparison between structures resulting from dimeric encounter complexes of WT and structures of the covalent disulphide-linked homodimer in the PDB structures (entries: 4R9H, 4RA3, 3TM6) is performed. The X-ray structures show an involvement of residues in BC loop and D strand, in agreement with docking results of WT (SI, Table ST1), indicating the importance of those regions in initial steps of oligomerization. A larger variability of dimeric interface in docking and MD simulations is expected due to the presence of a larger number of degrees of freedom involved in the association process between monomers. The three variants are protonated with the aid of the server H++ (http://biophysics.cs.vt.edu/H++) at a pH = 7.0 in order to be close to physiological conditions, resulting in a net charge of -2 for WT, +2 for D76N and -3 for ΔN6. To simulate a neutral system, we add the proper amount of Na or Cl ions for monomers and dimers of each species. Furthermore, the presence of the disulfide bridge between Cys25 and Cys80 is modeled by GROMACS.
4.1 Docking
A preliminary docking to build protein dimers is performed. Rigid-body docking method implemented in SDA 7.2 [29] are applied to predict dimeric interfaces of encounter complexes. During the simulations, translational and rotational motions are simulated for one protein with respect to the position of another identical protein. This procedure is performed individually for WT and each mutants. In SDA software, every trajectory begins with two proteins at center-to-center distance b, and stops when a protein reaches center-to-center distance c with a larger value than b. Here, b is set to 100 Å and c to 200 Å, and the temperature is set to 300 K. The reaction criterion defines the encounter event and here we consider a successful complex to be formed when two independent contacts come within 5.5 Å. In such procedure, performed individually for the WT and for each of the mutants, the contacts should be between interface residues predefined by SDA. The diffusion coefficients are set to 0.0135 Å2/ps and 3.92 10-5 radian2/ps for the relative translational coefficient and the rotational coefficient of each protein, respectively. The time step is set to 1.0 ps for center-to-center distances up to 50 Å, and for larger distances it increases linearly. For calculation of hydrophobic desolvation energy/forces it is considered a parameter value of -0.013 kcal/mol/Å2 and for the electrostatic desolvation energy/forces a parameter value of 1.67 kcal/mol/Å2, according to Elcock et al. [43]. Ionic strenght is set to 50 mM. We apply a single-linkage clustering method based on CA atoms and characterized by an RMSD of 3.0 Å in order to provide the smallest number of physically distinct orientations of one protein relative to the position of the other. All results given in the manuscript start from such configurations.
4.2 Molecular Dynamics (MD) Simulations
After performing initial docking, stability of encounter complexes is reached by running atomistic simulations for both dimers and monomers in water (the latter used as a benchmark) using GROMACS (v4.6.7)[44]. Simulations are based on the OPLS-AA force field with the SPC/E water model as implemented in the GROMACS package. The electrostatic potential, in its long-range part, is treated with particle-mesh-Ewald (PME) protocol. The radius for electrostatic interactions is set to 1.10 nm, while the long range radius is set to 1.00 nm. The simulations are performed using periodic boundary condition (PBC) in all directions. The bond lengths are treated like fixed constraints and a time step of 0.002 ps for numerical integration of the equations of motions is used. A simulation protocol chosen in our group’s previous works [47] is adopted: it includes an initial energy minimization using steepest descent algorithm, followed by first equilibration simulation of 500 ps under constant volume at 300 K (temperature coupling: velocity rescale [45]) and subsequently an equilibration simulation of 500 ps with a constant pressure of 1 bar (pressure coupling: Parrinello-Rahman [46]) is carried out. Finally, the stability of selected complexes is examined by running standard MD simulations at 300 K of a length of 200 ns for monomers and of 400 ns for dimers. Simulations are repeated four times using a different seed for starting velocity distribution to improve statistics. The trajectories are analyzed following a standard analysis and using tools within the GROMACS package.
4.3 Temperature Replica-exchange Simulations (T-REMD)
Due to the intrinsically limited time scale accessible to classical MD, the parallel tempering in the form of replica exchange molecular dynamics (REMD) is also performed, following previous protocol simulation of our group [48, 50]; in particular, such simulations are applied to dimers exhibiting the most extended dimeric interface (D76N and ΔN6). REMD involves multiple independent simulations at different temperatures (T-REMD), during which running replicas periodically attempt an exchange in temperature space [51, 53]. Replicas at low temperature have an opportunity to exchange to a higher temperature where energy barriers may be more easily crossed, enhancing the effective sampling space. In fact, one of the methodological advantages of T-REMD is that a protein can explore different folding pathways at the same time and can overcome barriers or trapped states at higher temperatures. For the investigated systems, 20 ns simulations on a total of 32 replicas are employed, covering the temperature range between 290 K and 320 K [54]. The exchange rate is 0.2 and the time interval each replica spends in a given temperature is 10 ps. Further details are reported in SI in Figure S11.
Acknowledgments
MCM, GB and SC acknowledge the CINECA award under the ISCRA initiative, for the availability of high performance computing resources and support. Oak Ridge National Laboratory is acknowledged for the supercomputing project CNMS2018-338, through the Scientific User Facilities Division, Office of Basic Energy Sciences, U.S. Department of Energy. Facilities of the National Energy Research Scientific Computing Center (NERSC), which is supported by the Office of Science of the U.S. Department of Energy under Contract No. DE-AC02-05CH11231, are also acknowledged.
References
[1] P. J. Bjorkman, M. A. Saper, B. Samraoui, W. S. Bennett, J. L. Strominger, and D. C. Wiley. Structure of the human class I histocompatibility antigen, HLA-A2. Nature, 329:506, 1987.
[2] L. Halabelian, S. Ricagno, S. Giorgetti, C. Santambrogio, A. Barbiroli, S. Pellegrino, A. Achour, R. Grandori, L. Marchese, S. Raimondi, P. P. Mangione, G. Esposito, R. Al-Shawi, J. P. Simons, I. Speck, M. Stoppini, M. Bolognesi, and V. Bellotti. Class I major histocompatibility complex, the trojan horse for secretion of amyloidogenic β2-microglobulin. Journal of Biological Chemistry, 289(6):3318–3327, 2014.
[3] F. Gejyo, T. Yamada, S. Odani, Y. Nakagawa, M. Arakawa, T. Kunitomo, H. Kataoka, M. Suzuki, Y. Hirasawa, T. Shirahama, A. S. Cohen, and K. Schmid. A new form of amyloid protein associated with chronic hemodialysis was identified as β2-microglobulin. Biochemical and Biophysical Research Communications, 129(3):701–706, 1985.
[4] A. Corazza, E. Rennella, P. Schanda, M. C. Mimmi, T. Cutuil, S. Raimondi, S. Giorgetti, F. Fogolari, P. Viglino, L. Frydman, M. Gal, V. Bellotti, B. Brutscher, and G. Esposito. Native-unlike long-lived intermediates along the folding pathway of the amyloidogenic protein β2-microglobulin revealed by real-time two-dimensional NMR. Journal of Biological Chemistry, 285(8):5827–5835, 2010.
[5] C. Rosano, S. Zuccotti, and M. Bolognesi. The three-dimensional structure of β2-microglobulin: results from x-ray crystallography. Biochimica et Biophysica Acta, 1753(1):85–91, 2005.
[6] M. Stoppini and V. Bellotti. Systemic amyloidosis: Lessons from β2-microglobulin. Journal of Biological Chemistry, 290(16):9951–9958, 2015.
[7] H. Naiki, N. Hashimoto, S. Suzuki, H. Kimura, K. Nakakuki, and F. Gejyo. Establishment of a kinetic model of dialysis-related amyloid fibril extension in vitro. Amyloid, 4(4):223–232, 1997.
[8] C. M. Eakin, A. J. Berman, and A. D. Miranker. A native to amyloidogenic transition regulated by a backbone trigger. Nature Structural Molecular Biology, 13:202, 2006.
[9] M. Stoppini, P. Mangione, M. Monti, S. Giorgetti, L. Marchese, P. Arcidiaco, L. Verga, S. Segagni, P. Pucci, G. Merlini, and V. Bellotti. Proteomics of β2-microglobulin amyloid fibrils. Biochimica et Biophysica Acta, 1753(1):23–33, 2005.
[10] G. Esposito, R. Michelutti, G. Verdone, P. Viglino, H. Hernández, C.V. Robinson, A. Amoresano, F. Dal Piaz, M. Monti, P. Pucci, P. Mangione, M. Stoppini, G. Merlini, G. Ferri, and V. Bellotti. Removal of the n-terminal hexapeptide from human β2-microglobulin facilities protein aggregation and fibril formation. Protein Science, 9(5):831–845, 2000.
[11] V. Bellotti, M. Gallieni, S. Giorgetti, and D. Brancaccio. Dynamic of β2-microglobulin fibril formation and reabsorption: The role of proteolysis. Seminars in Dialysis, 14(2):117–122, 2001.
[12] T. Eichner, A. P. Kalverda, G. S. Thompson, S. W. Homans, and S. E. Radford. Conformational conversion during amyloid formation at atomic resolution. Molecular Cell, 41(2):161–172, 2011.
[13] S. Valleix, J. D. Gillmore, F. Bridoux, P. P . Mangione, A. Dogan, B. Nedelec, M. Boimard, G. Touchard, J. M. Goujon, C. Lacombe, P. Lozeron, D. Adams, C. Lacroix, T. Maisonobe, V. Plante-Bordeneuve, J. A. Vrana, J. D. Theis, S. Giorgetti, R. Porcari, S. Ricagno, M. Bolognesi, M. Stoppini, M. Delpech, M. B. Pepys, P. N. Hawkins, and V. Bellotti. Hereditary systemic amyloidosis due to Asp76Asn variant β2-microglobulin. New England Journal of Medicine, 366(24): 2276–2283, 2012.
[14] M. de Rosa, A. Barbiroli, S. Giorgetti, P. P. Mangione, M. Bolognesi, and S. Ricagno. Decoding the structural bases of D76N β2-microglobulin high amyloidogenicity through crystallography and ASN-scan mutagenesis. PLosOne, 1–15, 2015.
[15] P. P. Mangione, G. Esposito, A. Relini, S. Raimondi, R. Porcari, S. Giorgetti, A. Corazza, F. Fogolari, A. Penco, Y. Goto, Y-H. Lee, H. Yagi, C. Cecconi, M. M. Naqvi, J. D. Gillmore, P. N. Hawkins, F. Chiti, R. Rolandi, G. W. Taylor, M. B. Pepys, M. Stoppini, and V. Bellotti. Structure, folding dynamics, and amyloidogenesis of D76N β2-microglobulin: roles of shear flow, hydrophobic surfaces and α-crystallin. Journal of Biological Chemistry, 288(43):30917–30930, 2013.
[16] K. Domanska, S. Vanderhaegen, V. Srinivasan, E. Pardon, F. Dupeux, J. A. Marquez, S. Giorgetti, M. Stoppini, L. Wyns, V. Bellotti, and J. Steyaert. Atomic structure of a nanobody-trapped domain-swapped dimer of an amyloidogenic β2-microglobulin variant. Proceedings of the National Academy of Sciences, 108(4):1314–1319, 2011.
[17] D. P. Smith, S. Jones, L. C. Serpell, M. Sunde, and S. E. Radford. A systematic investigation into the effect of protein destabilisation on β2-microglobulin amyloid formation. Journal of Molecular Biology, 330(5):943–954, 2003.
[18] T. R. Jahn, M. J. Parker, S. W. Homans, and S. E. Radford. Amyloid formation under physiological conditions proceeds via a native-like folding intermediate. Nature Structural and Molecular Biology, 13, 2006.
[19] T. Eichner and S. E. Radford. A generic mechanism of β2-microglobulin amyloid assembly at neutral pH involving a specific proline switch. Journal of Molecular Biology, 386(5):1312–1326, 2009.
[20] G. W. Platt and S. E. Radford. Glimpses of the molecular mechanisms of β2-microglobulin fibril formation in vitro: aggregation on a complex energy landscape. FEBS Letter, 583(16):2623–2629, 2009.
[21] N. H. H. Heegaard, T. J. D. Jørgensen, N. Rozlosnik, D. B. Corlin, J. S. Pedersen, A. G. Tempesta, P. Roepstorff, R. Bauer, and M. H. Nissen. Unfolding, aggregation, and seeded amyloid formation of Lysine-58-cleaved β2-microglobulin. Biochemistry, 44(11):4397–4407, 2005.
[22] S. Giorgetti, S. Raimondi, K. Pagano, A. Relini, M. Bucciantini, A. Corazza, F. Fogolari, L. Codutti, M. Salmona, P. Mangione, L. Colombo, A. De Luigi, R. Porcari, A. Gliozzi, M. Stefani, G. Esposito, V. Bellotti, and M. Stoppini. Effect of tetracyclines on the dynamics of formation and destructuration of β2-microglobulin amyloid fibrils. Journal of Biological Chemistry, 286(3):2121–2131, 2011.
[23] F. Fogolari, A. Corazza, P. Viglino, P. Zuccato, L. Pieri, P. Faccioli, V. Bellotti, and G. Esposito. Molecular dynamics simulation suggests possible interaction patterns at early steps of β2-microglobulin aggregation. Biophysical Journal, 92(5):1673–1681, 2007.
[24] S. G. Estacio, H. Krobath, D. Vila-Vicosa, M. Machuqueiro, E. I. Shakhnovich, and P. F. N. Faisca. A simulated intermediate state for folding and aggregation provides insights into ΔN6 β2-microglobulin amyloidogenic behavior. PLoS Computational Biology, 10(5):1–17, 2014.
[25] C. M. Eakin, F. J. Attenello, C. J. Morgan, and A. D. Miranker. Oligomeric assembly of native-like precursors precedes amyloid formation by β2-microglobulin. Biochemistry, 43(24):7808–7815, 2004.
[26] A. M. Smith, T. R. Jahn, A. E. Ashcroft, and S. E. Radford. Direct observation of oligomeric species formed in the early stages of amyloid fibril formation using electrospray ionisation mass spectrometry. Journal of Molecular Biology, 364(1):9–19, 2006.
[27] H. E. White, J. L. Hodgkinson, T. R. Jahn, S. Cohen-Krausz, W. S. Gosal, S. Müller, E. V. Orlova, S. E. Radford, and H. R. Saibil. Globular tetramers of β2-microglobulin assemble into elaborate amyloid fibrils. Journal of Molecular Biology, 389(1):48–57, 2009.
[28] E. Rennella, T. Cutuil, P. Schanda, I. Ayala, F. Gabel, V. Forge, A. Corazza, G. Esposito, and B. Brutscher. Oligomeric states along the folding pathways of β2-microglobulin: kinetics, thermodynamics, and structure. Journal of Molecular Biology, 425(15):2722–2736, 2013.
[29] M. Martinez, N. J. Bruce, J. Romanowska, D. B. Kokh, M. Ozboyaci, X. Yu, M. A. Öztürk, S. Richter, and R. C. Wade. SDA 7: a modular and parallel implementation of the simulation of diffusional association software. Journal of Computational Chemistry, 36(21):1631–1645, 2015.
[30] D. J. Earl and M. W. Deem. Parallel tempering: Theory, applications and new perspectives. Physical Chemistry Chemical Physics, 7:3910–3916, 2005.
[31] G. Verdone, A. Corazza, P. Viglino, F. Pettirossi, S. Giorgetti, P. P. Mangione, A. Andreola, M. Stoppini, V. Bellotti, and G. Esposito. The solution structure of human β2-microglobulin reveals the prodromes of its amyloid transition. Protein Science, 11(3):487–499, 2002.
[32] D. Gumral, F. Fogolari, A. Corazza, P. Viglino, S. Giorgetti, M. Stoppini, V. Bellotti, and G. Esposito. Reduction of conformational mobility and aggregation in W60G β2-microglobulin: assessment by N15 NMR relaxation. Magnetic Resonance in Chemistry, 51(12):795–807, 2015.
[33] G. Esposito, M. Garvey, V. Alverdi, F. Pettirossi, A. Corazza, F. Fogolari, M. Polano, P. P. Mangione, S. Giorgetti, M. Stoppini, A. Rekas, V. Bellotti, A.J.R. Heck and J. A. Carver. Monitoring the interaction between β2-microglobulin and the molecular chaperone αB-crystallin by NMR and mass spectrometry: αb-crystallin dissociates β2-microglobulin oligomers. The Journal of Biological Chemistry, 288(24):17844–17858, 2013.
[34] G. Brancolini, D. Toroz, and S. Corni. Can small hydrophobic gold nanoparticles inhibit β2-microglobulin fibrillation? Nanoscale, 6: 7903–7911, 2014.
[35] L. Halabelian, A. Relini, A. Barbiroli, A. Penco, M. Bolognesi, and S. Ricagno. A covalent homodimer probing early oligomers along amyloid aggregation. Scientific Reports, 5, 2015.
[36] M. Beerbaum, M. Ballaschk, N. Erdmann, C. Schnick, A. Diehl, B. Uchanska-Ziegler, A. Ziegler, and P. Schmieder. NMR spectroscopy reveals unexpected structural variation at the protein-protein interface in MHC class I molecules. Journal of Biomolecular NMR, 57(2):167–178, 2013.
[37] R. J. S. Loureiro, D. Vila-Vicosa, M. Machuqueiro, E. I. Shakhnovich, and P. F. N. Faisca. A tale of two tails: The importance of unstructured termini in the aggregation pathway of β2-microglobulin. Proteins: Structure, Function, and Bioinformatics, 85(11):2045–2057, 2017.
[38] S. Raimondi, R. Porcari, P. P. Mangione, G. Verona, J. Marcoux, S. Giorgetti, G. W. Taylor, S. Ellmerich, M. Ballico, S. Zanini, E. Pardon, R. Al-Shawi, J. P. Simons, A. Corazza, F. Fogolari, M. Leri, M. Stefani, M. Bucciantini, J. D. Gillmore, P. N. Hawkins, M. Valli, M. Stoppini, C. V. Robinson, J. Steyaert, G. Esposito, and V. Bellotti. A specific nanobody prevents amyloidogenesis of D76N β2-microglobulin in vitro and modifies its tissue distribution in vivo. Scientific Reports, 7:46711, 2017.
[39] A specific nanobody prevents amyloidogenesis of D76N The controlling roles of Trp60 and Trp95 in β2-microglobulin function, folding and amyloid aggregation properties. Journal of Molecular Biology, 378:887, 2008.
[40] V. L. Mendoza, K. Antwi, M. A. Barón-Rodríguez, C. Blanco, and R. W. Vachet. Structure of the preamyloid dimer of β2-microglobulin from covalent labeling and mass spectrometry. Biochemistry, 49(7): 1522–1532, 2010.
[41] G. W. Platt, K. E. Routledge, S. W. Homans, and S. E. Radford. Fibril growth kinetics reveal a region of β2-microglobulin important for nucleation and elongation of aggregation. Journal of Molecular Biology, 378(1):251–263, 2008.
[42] T. Eichner and S. E. Radford. Understanding the complex mechanisms of β2-microglobulin amyloid assembly. FEBS Journal, 278(20): 3868–3883, 2011.
[43] A. H. Elcock, R. R. Gabdoulline, R. C. Wade, and J. A. McCammon. Computer simulation of protein-protein association kinetics: acetylcholinesterase-fasciculin. Journal of Molecular Biology, 291(1):149–162, 1999.
[44] D. Van Der Spoel, E. Lindahl, B. Hess, G. Groenhof, A. E. Mark, and H. J. C. Berendsen. Gromacs: Fast, flexible, and free. Journal of Computational Chemistry, 26(16):1701–1718, 2005.
[45] G. Bussi, D. Donadio, and M. Parrinello. Canonical sampling through velocity rescaling. Journal of Chemical Physics, 126(1), 2007.
[46] M. Parrinello and A. Rahman. Polymorphic transitions in single crystals: A new molecular dynamics method. Journal of Applied Physics, 52(12):7182–7190, 1981.
[47] G. Brancolini, D. B. Kokh, L. Calzolai, R. C. Wade, and S. Corni. Docking of ubiquitin to gold nanoparticles. ACS Nano, 6(11):9863–9878, 2012.
[48] G. Brancolini, A. Corazza, M. Vuano, F. Fogolari, M. C. Mimmi, V. Bellotti, M. Stoppini, S. Corni, and G. Esposito. Probing the influence of citrate-capped gold nanoparticles on an amyloidogenic protein. ACS Nano, 9(3):2600–2613, 2015.
[49] C. Cantarutti, S. Raimondi, G. Brancolini, A. Corazza, S. Giorgetti, M. Ballico, S. Zanini, G. Palmisano, P. Bertoncin, L. Marchese, P. P. Mangione, V. Bellotti, S. Corni, F. Fogolari, and G. Esposito. Citrate-stabilized gold nanoparticles hinder fibrillogenesis of a pathological variant of β2-microglobulin. Nanoscale, 9:3941–3951, 2017.
[50] G. Brancolini, M. C. Maschio, C. Cantarutti, A. Corazza, F. Fogolari, V. Bellotti, S. Corni, and G. Esposito. Citrate stabilized gold nanoparticles interfere with amyloid fibril formation: D76N and δN6 β2-microglobulin variants. Nanoscale, 10:4793–4806, 2018.
[51] A. Patriksson and D. van der Spoel. A temperature predictor for parallel tempering simulations. Physical Chemistry Chemical Physics, 10:2073–2077, 2008.
[52] M. M. Seibert, A. Patriksson, B. Hess, and D. van der Spoel. Reproducible polypeptide folding and structure prediction using molecular dynamics simulations. Journal of Molecular Biology, 354(1):173–183, 2005.
[53] D. van der Spoel and M. M. Seibert. Protein folding kinetics and thermodynamics from atomistic simulations. Physical Review Letter., 96:238102, 2006.
[54] S. Kirkpatrick, C. D. Gelatt, and M. P. Vecchi. Optimization by simulated annealing. Science, 220(4598):671–680, 1983.
Biographies

Maria Celeste Maschio is a PhD student at the University of Modena and Reggio Emilia, Italy. She is working on a project about the modeling of an amyloidogenic protein in solution and on nanostructures, using classical molecular dynamics, under the supervision of Prof. Stefano Corni and Giorgia Brancolini. She spent a period of six months in King’s College London (UK) working with Prof. Carla Molteni on enhanced sampling methods as Metadynamics.

Giorgia Brancolini received her Ph.D. in Chemistry from the University of Modena and Reggio Emilia, Italy, in cooperation with the University of Ulm, Germany. She was a post-doctoral fellow at Politecnico di Milano from 2002–2004 in the Dept. of Chemical Engineering. From 2004–2010 she was a senior post-doctoral fellow at National Research Council in Italy. Since 2010 Dr. Brancolini is a fixed term research scientist at the Center S3 CNR-NANO of the Institute of Nanoscience of the National Research Council in Modena, Italy. She is working on multi scale simulations, spanning from ab initio to classical and coarse grained models, of the interactions between proteins and inorganic surfaces/nanoparticles.

Stefano Corni is full-professor of Physical Chemistry at the Dept. of Chemical Sciences, University of Padova, Italy, and a research associate with CNR-NANO Modena, Italy. He got his Ph.D. in Chemistry from the Scuola Normale Superiore di Pisa, Italy, in 2003. He is working on the classical and quantum mechanical modeling of the interactions between proteins and inorganic surfaces/nanoparticles, on the optical properties of molecules interacting with metal nanoparticles and on electron transfer in biomolecules and between biomolecules and electrodes.
Journal of Self-Assembly and Molecular Electronics, Vol. 6_1, 35–60.
doi: 10.13052/jsame2245-4551.6.003
This is an Open Access publication. © 2019 the Author(s). All rights reserved.