Very Fast Algorithms for Eliminating the Diffraction Effects in Protein-Based Volumetric Memories
Received 12 December 2014; Accepted 17 February 2015; Publication 31 March 2015
Journal of Self-Assembly and Molecular Electronics, Vol. 2, 41ȁ52.
doi: 10.13052/jsame2245-4551.213
© 2015 River Publishers. All rights reserved.
Dragos Trinca1 and Sanguthevar Rajasekaran2
- 1Sc Piretus Prod Srl, Osoi, Jud. Iasi, 707110, Romania dntrinca@gmail.com
- 2Dep. of Computer Science and Engineering, University of Connecticut, Storrs, USA rajasek@engr.uconn.edu
Abstract
One of the current research directions in biological nanotechnology is the use of bacteriorhodopsin in the fabrication of protein-based volumetric memories. Bacteriorhodopsin, with its unique light-activated photocycle, nanoscale size, cyclicity (>107), and natural resistance to harsh environmental conditions, provides for protein-based volumetric memories that have a comparative advantage over magnetic and optical data storage devices. The construction of protein-based volumetric memories has been, however, severely limited by fundamental issues that exist with such devices, such as unwanted diffraction effects. In this paper, we propose some optimizations that can be applied to one of the previously proposed algorithms for eliminating the diffraction effects.
Keywords
- compression
- biological nanotechnology
- optimization
1 Introduction
Much of the current research effort in biological nanotechnology is directed toward self-assembled monolayers and thin films, biosensors, and protein-based photonic devices [1–6]. Although a number of proteins have been explored for device applications, bacteriorhodopsin [3] has received, it seems, the most attention. Bacteriorhodopsin protein memory devices exhibit increased thermal, chemical and photochromic stability, and have the advantage of being radiation-hardened, waterproof, and EMP-resistant. Such devices are capable of storing large amounts of data in a small volume.
Although there are a number of prototype systems and preliminary effort to apply them, the potential of this promising technology is relatively unexplored. Research on protein-based memories started in the late 1980s with considerable anticipation, but enthusiasm decreased shortly afterwards for several reasons. Commercial development of spatial light modulators (SLMs), that are an integral component of protein-based memories, was slow and there were several issues, such as unwanted diffraction effects, that limited performance in three-dimensional memory applications. More recently, however, the development of high-definition television projection equipment has resulted in the commercial availability of high-resolution, high-performance and relatively inexpensive SLMs. But, fundamental problems have remained. Two such problems are diffraction effects and scaling.
1.1 Diffraction Effects
The branched-photocycle three-dimensional memory, as discussed in [2, 7], stores data by using a sequential two-photon process to convert bacteriorhodopsin (bR) in the activated region from the bR resting state to the Q state. This process involves using a paging beam to select a thin page of memory and a writing beam that is pixilated at those positions where data are to be written. The transition from the bR resting state to the Q state occurs via the intermediate states K, L, M, N, and O. Only the bR (bit 0) and Q (bit 1) states are stable for long periods of time. By using 32-level grey-scaling and two polarizations, each voxel can store 64 bits. Attempts to use higher levels of grey-scaling have failed, mainly due to the problems of diffraction introduced by having pages with significant differences in the average refractive indices. To understand this problem, we note that protein corresponding to bit 0 has a high refractive index and protein corresponding to bit 1 has a low refractive index with reference to the red laser beam at 633 nm that is used to read out the data. Prototypes made of the three-dimensional memory fail to work at high storage densities when individual pages of memory have a preponderance of bits of a given state. Consider the worst case – each page is either all 0’s or all 1’s. Then, a refractive index grating is created, that diffracts the laser beams quite efficiently because individual pages are stored with separations of 6-20 µm. While these separations are significantly larger than the diffraction limit would dictate, closer spacing is not possible, due to beam steering inside the data cuvettes. The unwanted beam steering is due to refractive index gradients. Algorithms that can store data at high resolution and that maintain the number of 0’s equal to the number of 1’s would solve this problem [7–12].
2 Optimizing the APPROXv3 Algorithm
One way of ensuring an equal number of 0’s and 1’s is to replace a 0 with 01 and a 1 with 10. However, this procedure would reduce the available memory by a factor of 2 (which means that the utility factor would be 50% in this case). In [7] and [9] the authors have proposed some methods that provide utility factors of more than 50%. For completely random data, however, the best utility factor provided by the previously proposed algorithms is about 99.9%, and can be obtained by using the APPROXv1 algorithm proposed in [9], which works as follows. Let I be the input data, and bI the binary representation of I. If I is of length L, then bI is of length 8L, as each byte of data is represented binary on 8 bits. Scan bI and count the number of 0’s and 1’s. Suppose that bI doesn’t have an equal number of 0’s and 1’s. Scan bI from left to right, bit by bit. For each position i in the scanning, 1 ≤ i ≤ 8L, do as follows. Let bI1:i be the portion already processed, and bIi+1:8L the portion that has remained to be processed. Let be the complement of bI1:i. If the binary string has an equal number of 0’s and 1’s, then stop the scanning, let q be this first position i that leads to a with an equal number of 0’s and 1’s, and the output of APPROXv1 is , where x is a binary string of length 256 such that the first 120 bits of x store q, the next 3 bits store the version of the algorithm (in this case 001), the next 5 bits are 00000 (not used in APPROXv1) and the remaining 128 bits are used to make x a binary string with an equal number of 0’s and 1’s. So, in conclusion, the length of the output is 8L+256. (There is no particular reason for choosing the length of x to be 256, just that this value seems to be sufficient in practice.)
In [11], some optimizations that can be applied to APPROXv1 have been proposed. Precisely, the optimizations proposed in [11] apply to the second scanning step (when the position q is found), and work as follows. Suppose that bI has more 0’s than 1’s. Let NofZs be the number of 0’s in bI, and NofOs the number of 1’s in bI. Observe that every position i that leads to a with an equal number of 0’s and 1’s has the property that bI1:i has exactly (i-((NofZs – NofOs)/2))/2+(NofZs – NofOs)/2 0’s and (i – ((NofZs – NofOs)/2))/2 1’s. So, during the scanning, we can skip those positions that for sure don’t have this property, and examine only those positions that may have this property, until the desired position is found. Precisely, the modifications are the following.
- First, we don’t have to start the scanning from position 1. We can start directly with position (NofZs – NofOs)/2, the reason being that we have to complement exactly (NofZs – NofOs)/2 0’s in order to transform bI into a binary string with an equal number of 0’s and 1’s.
- Second, let i be the current position in the scanning, and let Zs[i] be the number of 0’s in bI1:i. The number of 1’s in bI1:i is, clearly, i – Zs[i]. We have two cases, either Zs[i] ≥ (i – Zs[i]) or Zs[i] < (i – Zs[i]).
- If , we check to see if . If yes, then the scanning stops, and let q be this first position i that leads to a with an equal number of 0’s and 1’s. Otherwise, if , then the next position in the scanning is .
- If , the next position in the scanning is .
This is the APPROXv2 algorithm recently proposed in [11]. The output in APPROXv2 is , where the first 120 bits of x store q, the next 3 bits are 010 (the version of the algorithm, which is 2), the next 5 bits are 00000 (also, not used in APPROXv2), and the remaining 128 bits are used to make x a binary string with an equal number of 0’s and 1’s, as usual.
Example 1 Suppose that
bI has 18 0’s and 14 1’s. APPROXv1 works as follows.
- Since has 17 0’s and 15 1’s, the scanning goes on to the next position.
- Since has 18 0’s and 14 1’s, the scanning goes on to the next position.
- Since has 17 0’s and 15 1’s, the scanning goes on to the next position.
- Since has 18 0’s and 14 1’s, the scanning goes on to the next position.
- Since has 19 0’s and 13 1’s, the scanning goes on to the next position.
- Since has 18 0’s and 14 1’s, the scanning goes on to the next position.
- Since has 19 0’s and 13 1’s, the scanning goes on to the next position.
- Since has 18 0’s and 14 1’s, the scanning goes on to the next position.
- Since has 19 0’s and 13 1’s, the scanning goes on to the next position.
- Since has 20 0’s and 12 1’s, the scanning goes on to the next position.
- Since has 19 0’s and 13 1’s, the scanning goes on to the next position.
- Since has 18 0’s and 14 1’s, the scanning goes on to the next position.
- Since has 17 0’s and 15 1’s, the scanning goes on to the next position.
- Since has 16 0’s and 16 1’s, we conclude that $q=14$, and the scanning stops.
Example 2 For the same bI, APPROXv2 works as follows.
- Since we have Zs [2] = 1 and 2 – Zs[2] = 1, the next position in the scanning is 4.
- Since we have Zs [4] = 2 and 4 – Zs[4] = 2, the next position in the scanning is 6.
- Since we have Zs [6] = 3 and 6 – Zs[6] = 3, the next position in the scanning is 8.
- Since we have Zs [8] = 4 and 8 – Zs[8] = 4, the next position in the scanning is 10.
- Since Zs [10] = 4 and 10 – Zs[10] = 6, the next position in the scanning is 14.
- Since Zs [14] = 8 and 14 – Zs[14] = 6 and Zs[14] – (14 – Zs[14]) = 2, we conclude that q=14.
- So, significantly less positions are checked in APPROXv2 as compared to APPROXv1.
APPROXv2 can be optimized as follows. Assume that, besides the input data, we have a parameter sl, such that 1 ≤ sl ≤ 8L, and such that 8L is a multiple of sl. Then, we consider the following substrings of bI:
So, we are splitting bI into substrings, each of length sl. For each , consider the pairs (ai, bi) and (ci, di), where
So, for each and sbi are the differences between the number of 0’s and 1’s at the two ends, while ci and di are the differences between the number of 1’s and 0’s at the two ends. APPROXv1 and APPROXv2 start with a scanning of bI where we count the number of 0’s and 1’s. The pairs (ai, bi), (ci, di) can be computed during this first scanning. We have the following [12]
Fact 1 If NofZs > NofOs, then there exists at least one i, , such that
Otherwise, if NofOs > NofZs, then there exists at least one i, , such that
Given this, APPROXv2 can be optimized as follows. Suppose that NofZs > NofOs. Let j be the first index that satisfies . Then, we can search for the position q only in the substring , because, at this point, we know that the position q we are looking for satisfies . This is the APPROXv3 algorithm, proposed in [12]. The output in APPROXv3 is as in APPROXv2, the only difference being x121:123 = 011 (the version of the algorithm, which is 3).
Example 3 For the same bI, and assumming sl = 8, APPROXv3 works as follows.
- First, we compute the pairs: (a1 = 1, b1 = 0), (a2 = -1, b2 = 2), (a3 = 1, b3 = 4), (a4 = 5, b4 = 4).
- The first pair (ai, bi) that satisfies
is (a2, b2).
- So, we search for the position q only in the substring bI9:16, and we find q = 14.
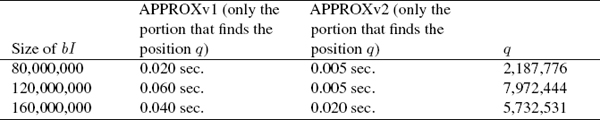
Some results when comparing APPROXv1 with APPROXv2 are given in Table 1. APPROXv3 takes less than one millisecond, in general.
Table 1 Experimental results
In this paper, we propose some optimizations that can be applied to APPROXv3, and that could lead to faster practical algorithms for eliminating the diffraction effects. Suppose that NofZs > NofOs, and let j be the first index that satisfies
Then, we search for the position q in the substring , and we find it.
If , then the output is , where x is a binary string of length 256 such that the first 120 bits of x store q, the next 3 bits are 100, the next 5 bits are 00000, and the remaining 128 bits are used to make x a binary string with an equal number of 0’s and 1’s. If , then the output is , where x is a binary string of length 256 such that the first 120 bits of x store q, the next 3 bits are 100, the next 5 bits are 00000, and the remaining 128 bits are used to make x a binary string with an equal number of 0’s and 1’s.
Denote this version of APPROXv3 by APPROXv4. Clearly, in the case , APPROXv4 is basically APPROXv3. But, when , we compute in the output of APPROXv4 the complement of bIq+1:8L instead of the complement of bI1:q, the reason being that in this case the length of bIq+1:8L is strictly smaller than the length of bI1:q. In practice, in the case , this means that the larger the difference is, the faster APPROXv4 is as compared to APPROXv3.
Example 4 Suppose that
bI has 18 0’s and 14 1’s. Assumming sl = 8, APPROXv4 works as follows.
- First, we compute the pairs: (a1 = 1, b1 = 0), (a2 = 1, b2 = 0), (a3 = 1, b3 = 2), (a4 = 3, b4 = 4).
- The first pair (ai, bi) that satisfies
is (a3, b3).
- Thus, we search for the position q only in the substring bI17:24, and we find q = 22.
- So, we are in the second case, when , that is, q is in the second half of bI.
- The output is , where x is a binary string of length 256 such that the first 120 bits of x store q = 22, the next 3 bits are 100, the next 5 bits are 00000, and the remaining 128 bits are used to make x a binary string with an equal number of 0’s and 1’s.
APPROXv4 can be further optimized as follows. Suppose that NofZs > NofOs, and let j be the first index that satisfies
Then, we search for the position q in the substring , and find it.
If , then the output is , where x is a binary string of length 256 such that the first 120 bits of x store q, the next 3 bits are 101, the next 5 bits are 00000, and the remaining 128 bits are used to make x a binary string with an equal number of 0’s and 1’s. If , then consider the input rI, where rIi = bI8L-i+1, that is, rI is the reverse of bI. For rI, we know that NofZs > NofOs. From the pairs (ai, bi) corresponding to bI, we consider the corresponding pairs for rI, say . Let j′ be the first index that satisfies
Then, we search for the position q′ in the substring , and find it. That is, q′ is the first position i in rI such that has an equal number of 0’s and 1’s. Then, the output is , where x is a binary string of length 256 such that the first 120 bits of x store q′, the next 3 bits are 101, the next 5 bits are 10000, and the remaining 128 bits are used to make x a binary string with an equal number of 0’s and 1’s. So, in the case , the first 120 bits of x store the position q′ instead of q and since then the 124-th bit of x, which is 1, will tell us, at decompression, that at compression the input rI has been considered instead of bI.
Denote this version of APPROXv4 by APPROXv5. In the case , APPROXv5 is basically APPROXv4. When , then in APPROXv5 we consider the input rI instead of bI. In practice, in the case , if q′ is significantly smaller than 8L – q (the length of then complementing rI1:q' instead of bIq+1:8L could lead to a significant speed-up.
We note that, in the case , APPROXv5 does additional work as compared to APPROXv4, by finding the position q′. However, this additional work would be negligible in general. In practice, if the decompression time is more important than the compression time, that is, if the time taken when decompressing the output is more important than the time taken when constructing it, then if q′ is significantly smaller than 8L – q, this means that decomplementing instead of would take signifincatly less time at decompression.
Example 5 Suppose that
as before. bI has 18 0’s and 14 1’s. Assumming sl = 8, APPROXv5 works as follows.
- First, we compute the pairs: (a1 = 1, b1 = 0), (a2 = 1, b2 = 0), (a3 = 1, b3 = 2), (a4 = 3, b4 = 4).
- The first pair (ai, bi) that satisfies
is (a3, b3).
- Thus, we search for the position q only in the substring bI17:24, and we find q = 22.
- So, we are in the second case, when , that is, q is the second half of bI.
- Thus, we consider
the reverse of bI.
- The pairs corresponding to rI would be , , , .
- The first pair that satisfies
is .
- Thus, we search for the position q′ only in the substring rI1:8, and we find q′ = 4.
- The output is , where x is a binary string of length 256 such that the first 120 bits of x store q′ = 4, the next 3 bits are 101, the next 5 bits are 10000, and the remaining 128 bits are used to make x a binary string with an equal number of 0’s and 1’s.
Similarly as in the case of APPROXv3, for inputs bI of length a few millions bits, both APPROXv4 and APPROXv5 take in our implementations around one second.
3 Discussion and Conclusion
The algorithms presented are indeed very fast in practice, which should provide for a real-time and fast working of protein-based memories. This conclusion comes from the fact that, for example, APPROXv5 takes around one second for an input of a few millions bits, so for a protein-based memory page of one thousand bits in size, APPROXv5 would normally take around one millisecond, which is quite fast. We anticipate that the processor latency caused by the execution of one of these algorithms for the correct compression (when writing a page in the protein-based memory) or the correct decompression (when reading a page from the protein-based memory) would be negligible, especially with the current technology, which means that in practice the protein-based memories should be quite fast.
References
[1] R. R. Birge, Photophysics and molecular electronic applications of the rhodopsins, Annual Review of Physical Chemistry, vol. 41, pp. 683–733 (1990)
[2] R. R. Birge, N. B. Gillespie, E. W. Izaguirre, A. Kusnetzow, A. F. Lawrence, D. Singh, Q. W. Song, E. Schmidt, J. A. Stuart, S. Seetharaman and K. J. Wise, Biomolecular electronics: protein-based associative processors and volumetric memories, Journal of Physical Chemistry B, vol. 103, pp. 10746–10766 (1999)
[3] N. A. Hampp, Bacteriorhodopsin: mutating a biomaterial into an optoelectronic material, Applied Microbiology and Biotechnology, vol. 53, pp. 633–639 (2000)
[4] J. R. Hillebrecht, J. F. Koscielecki, K. J. Wise, D. L. Marcy, W. Tetley, R. Rangarajan, J. Sullivan, M. Brideau, M. P. Krebs, J. A. Stuart and R. R. Birge, Optimization of protein-based volumetric optical memories and associative processors by using directed evolution, NanoBiotechnology, vol. 1, pp. 141–151 (2005)
[5] D. Marcy, W. Tetley, J. Stuart and R. Birge, Three-dimensional data storage using the photochromic protein bacteriorhodopsin, in Proc. of the 22nd Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC'00), pp. 1003–1006
[6] D. Oesterhelt, C. Bräuchle and N. Hampp, Bacteriorhodopsin: a biological material for information processing, Quarterly Reviews of Biophysics, vol. 24, pp. 425–478 (1991)
[7] S. Rajasekaran, V. Kumar, S. Sahni and R. Birge, Efficient algorithms for protein-based associative processors and volumetric memories, in Proc. of IEEE NANO 2008, pp. 397–400
[8] S. Rajasekaran, V. Kundeti, R. Birge, V. Kumar and S. Sahni, Efficient algorithms for computing with protein-based volumetric memory processors, IEEE Transactions on Nanotechnology, vol. 10, pp. 881–890 (2010)
[9] D. Trincă and S. Rajasekaran, Coping with diffraction effects in protein-based computing through a specialized approximation algorithm with constant overhead, in Proc. of IEEE NANO 2010, pp. 802–805
[10] D. Trincă and S. Rajasekaran, Specialized compression for coping with diffraction effects in protein-based volumetric memories: solving some challenging instances, Journal of Nanoelectronics and Optoelectronics, vol. 5, pp. 290–294 (2010)
[11] D. Trincă and S. Rajasekaran, Optimizing the APPROXv1 algorithm for coping with diffraction effects in protein-based volumetric memories, in Technical Summaries of SPIE Optical Systems Design, abstract 8167A-71 (at page 14), 2011
[12] D. Trincă and S. Rajasekaran, Fast Algorithms for Coping with Diffraction Effects in Protein-based Volumetric Memories (Design and Implementation), Journal of Computational and Theoretical Nanoscience, vol. 10, pp. 894–897 (2013)
Biographies
D. Trinca received his BSc degree in 2003, from Facultatea de Informatica, Universitatea Alexandru Ioan Cuza din Iasi, Romania, and his PhD degree in 2008, from the Department of Computer Science and Engineering, University of Connecticut, USA. Currently he is with Sc Piretus Prod Srl in Osoi, jud. iasi, Romania. His research interests are in the area of computational problems in nanotechnology.
S. Rajasekaran received his BSc degree in Physics in 1977, from Madurai Kamaraj University, India, and his PhD degree in Computer Science in 1988, from Harvard University, USA. Currently he is with University of Connecticut, USA. His research interests are in the area of applied algorithms.