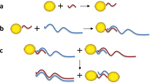
Formation of Dimers Composed of a Single Short dsDNA Connecting Two Gold Nanoparticles
Received 27 November 2012; Accepted 17 December 2012
Copyright © 2013 River Publishers. All rights reserved.
Haya Dachlika1, Avigail Stern1, Dvir Rotem1, Danny Porath*
- 1These authors made equal contributions to this manuscript.
Institute of Chemistry and The Harvey M. Krueger Center for Nanoscience and Nanotechnology, The Hebrew University of Jerusalem, 91904 Jerusalem, Israel - 2Corresponding author: danny.porath@mail.huji.ac.il
Abstract
We report synthesis of dimers composed of a single short double-stranded (ds)DNA molecule connecting two gold nanoparticles (GNPs). Such structures may be useful for electrical transport measurements through dsDNA molecules and for other research purposes. When the DNA molecules are short with respect to the size of the GNP, gel electrophoresis cannot separate GNPs with different numbers of DNA molecules attached to them. We present two methods to separate GNPs connected to single short thiolated single-stranded (ss)DNA. The separation is performed by hybridizing the DNA/GNP conjugates with long, partially complementary, ssDNA or with complementary ssDNA connected to GNPs of smaller size. The separated GNPs with a single short ssDNA were used to form dimers consisting of GNPs connected by a single short dsDNA molecule.
Keywords
- DNA
- gold nanoparticles
- gel electrophoresis
1. Introduction
Many conjugate structures composed of DNA and gold nanoparticles (GNPs) have been reported in the literature. A common method for forming these conjugates is by connecting the DNA strands to the GNP through thiol chemistry, which offers the ability to design complex nanoparticle assemblies [1–3]. Such conjugates have been used for various emerging challenges such as electrical transport measurements [4–6], surface plasmon resonance (SPR) studies [7, 8], molecular nanomachines formation [9] and others. Thanks to their optical properties [10], DNA/GNP conjugates were also applied to the development of applications in biosensing [11, 12], nanobiotechnology [11], nanomedicine [13, 14] and nanoelectronics [15]. For many of these applications it is important to control the exact number of DNA strands attached to the nanoparticles. The ability to separate conjugates of GNPs with a specific number of short DNA strands, as building blocks for 2D and 3D GNP-DNA nano-constructions, e.g. dimers, is challenging and has attracted much scientific interest.
Gel electrophoresis is a common technique for separation of DNA/GNP conjugates because additional DNA molecules connected to the GNP produce a significant shift in the electrophoretic mobility of the conjugates [16, 17]. Gel electrophoresis separation is, however, limited to particles connected to relatively long oligonucleotide strands. The sensitivity and the effectiveness of the method are reduced when the particle size is comparable or larger than the DNA length. Recently, some methods to synthesize GNPs attached to specific numbers of DNA strands have been employed: long DNA strands were first used to get efficient separation with gel electrophoresis and then removed or shortened in order to get GNPs with short DNA strands [18, 19]. Another method used anion-exchange HPLC as a technique for conjugate separation [20].
In this work we present two easy and convenient methods for separation of conjugates of GNPs with specific numbers of short oligonucleotide strands. The separation is implemented for and confirmed by the formation of GNP dimers connected by single dsDNA molecules. The separation methods are based on intermediate increase in the variability between the conjugates. This leads to a significant difference in the electrophoretic mobility of the different structures. Characterization of the products was done by transmission electron microscopy (TEM).
Table 1 DNA sequences used in the synthesis of the four DNA/GNP conjugates.
2. Experimental Procedures
2.1. Generation of GNP/DNA Conjugates
Citrate stabilized GNPs with mean diameters of 5 and 8 nm were purchased (Ted Pella). The GNPs were coated with phosphine ligands (bis(p-sulfonatophenyl)-phenylphosphine (BSPP) as previously described by Mastroianni et al. [21]. The GNPs were stabilized with 1 mM BSPP by overnight incubation at room temperature. NaCl was then added to the GNP solution until a color change from red to blue was observed (the final NaCl concentration was ∼200 mM for 8 nm GNP and ∼300 mM for 5 nm GNP). The GNP solution was concentrated by centrifugation at 3000 rpm for 10 minutes at room temperature. The supernatant was removed and the pellet was resuspended in 1 mg ml−1 BSPP solution (concentration of 3.14 mM) to ∼1 μM. The phosphine coating provides a net negative charge on the GNP surface, thus stabilizing it in high concentrations and in buffer conditions.
5’ thiol-C6-modified ssDNA oligonucleotides with 26 bases (sequences presented in Table 1) were purchased (Syntezza). The oligonucleotides were treated with tris(2-carboxyethyl)phosphine hydrochloride (TCEP) at a TCEP:DNA molar ratio of 1000:1 for removal of the protective group on the thiol, and then purified with Bio-Spin 6 columns (Biorad).
GNP/DNA conjugates were formed by incubating the BSPP coated GNPs with the treated DNA at a final concentration of 0.5 μM, DNA:GNP molar ratio of 1:1 overnight at room temperature (25°C) in 100 mM tris buffer (pH 7.4) with 50 mM NaCl.
2.2. Gel Electrophoresis Separation and Characterization
Two methods were used to separate conjugates of 8 nm GNPs with different numbers of 26 bases ssDNA strands. In the first method the DNA strands were conjugated to 8 nm GNPs and hybridized to complementary 26 bases ssDNA strands, which were conjugated to 5 nm GNPs. For the hybridization the two conjugate solutions (5 nm GNP conjugates and 8 nm GNP conjugates) were mixed at a 10:1 molar ratio, respectively, and incubated over night at room temperature. After the hybridization, the obtained structures were separated by gel electrophoresis in 3% MetaPhor agarose (Lonza) with 1X TBE as running buffer at 100 V for 60 min.
In the second method 100 bases ssDNA strands were hybridized to 26 bases ssDNA strands (over 26 bp length), which were already conjugated to the GNPs using thiols. The molar ratio of the hybridized 26bp:100bp conjugates was 1:10 respectively and the incubation was performed overnight at room temperature. The resulting structures were separated by gel electrophoresis in a 3% MetaPhor agarose gel with 1X Tris/Borate/EDTA (TBE) buffer as running buffer for 1.5 h at 100 V. The separation of the 20 nm GNPs was done in a 1.5% MetaPhor agarose gel under the same conditions.
2.3. Dimers Preparation
Conjugates of GNPs with DNA molecules that were prepared by the second method described above were extracted from the gel by electroelution at 100 V for half an hour. Conjugates of GNPs bound to a single ssDNA molecule (mono-conjugates) were concentrated similarly to the concentration procedure described above for BSPP stabilized GNPs. Mono-conjugates with complementary DNA sequences were then mixed, heated to 60°C for 10 min (for dehybridization of the 100 bases strands from the 26 bases strands and their hybridization with the complementary 100 bases strands) and then cooled down to room temperature (for hybridization of the 26 bases strands to the complementary ones). Hybridization of complementary 100 bases strands and complementary 26 bases strands is thermodynamically favored. Following hybridization of the complementary mono-conjugates, the structures were again separated by 3% MetaPhor agarose gel electrophoresis with 1X TBE as the running buffer at 100 V for 60 min. Dimers were then extracted from the gel by electroelution at 100 V for 60 min.
As a control experiment, dimers were also prepared without using any separation method. These dimers were prepared by mixing non-separated 8 nm GNP/DNA conjugate solutions and incubating them over night at room temperature for hybridization.
2.4. Deposition and TEM Imaging
Synthesized dimers were deposited on a 400 square mesh carbon coated copper TEM grid (Electron Microscopy Sciences) and imaged by a Tecnai T12 G2 Spirit TEM (FEI). Deposition was carried out by first treating the grid with 0.1% w/v poly-L-lysine in water for 1 min, washing by floating on a water drop for another minute, and then floating on the sample drop for three minutes. Images were taken with an FEI Eagle 4k, 16 Megapixel CCD camera.
3. Results and Discussion
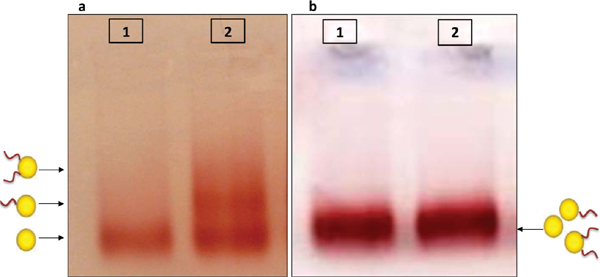
Gel electrophoresis is often used as a tool in the characterization and purification of GNPs/DNA conjugates [16, 17]. The basic principle relies on the characteristic mobility of objects in a porous matrix, or gel, under an electric field. The mobility (distance/(electric field * time)) depends mainly on the size and the charge of the conjugates [16]. A good separation between GNPs bound to different numbers of DNA molecules is achieved when the bound DNA molecules are much longer than the GNP diameter, e.g., GNPs with a diameter of 8 nm and ssDNA of 86 bases (see Figure 1(a)) [16, 20]. When the DNA molecules are short relative to the size of the GNPs, it is difficult or even impossible to separate DNA/GNP conjugates based on the number of bound DNA molecules. For instance, in the gel shown in Figure 1(b) it is impossible to distinguish between bare GNPs (lane 1) and conjugates of 8 nm GNPs bound to short, 26 bases long, thiolated DNA oligonucleotides (lane 2). Therefore, in order to prepare mono-conjugates of 8 nm GNP with a single 26 bases ssDNA molecule more refined separation methods must be used.
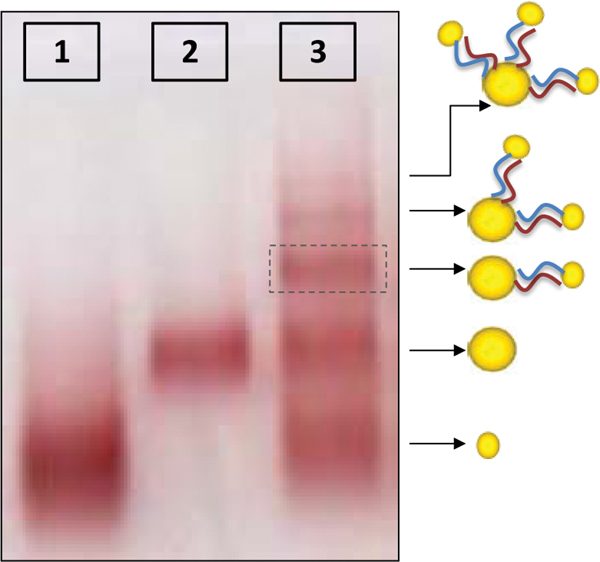
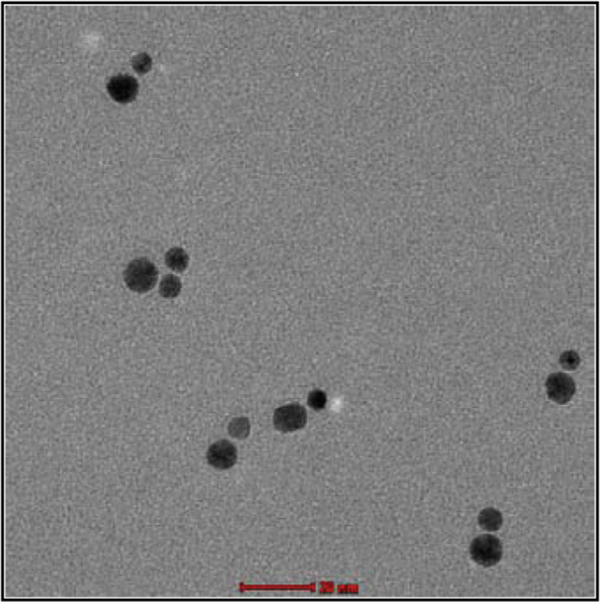
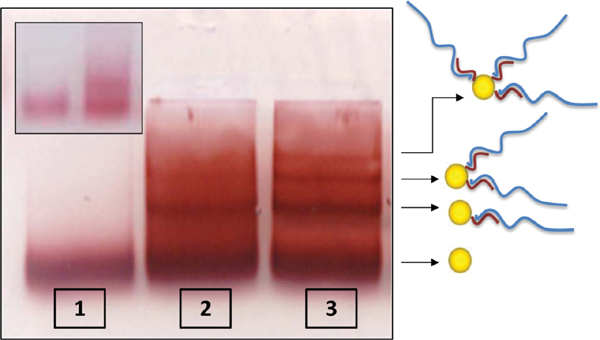
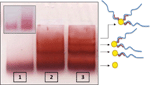
We describe two methods that enable to separate GNPs connected to different numbers of short ssDNA strands. These methods are based on temporary hybridization of the short DNA strands that are bound to the GNPs to complementary short ssDNA bound to a smaller GNP or to longer ssDNA strands. In the first method 8 nm GNPs were first bound to 26 bases ssDNA molecules (strand S26 1) and 5 nm GNPs were bound to the complementary 26 bases ssDNA molecules (strand S26 2). Then, the two solutions of GNP/DNA conjugates were mixed, hybridized, (with the 5 nm conjugates in a large excess) and separated by gel electrophoresis (Figure 2). Because the small conjugates were in large excess, they were assumed to hybridize with all the DNA strands bound to the 8 nm GNPs. This way the 5 nm GNP conjugates enable separation of the 8 nm GNP conjugates based on the number of DNA molecules (and 5 nm GNPs) bound to them. Therefore, after separation we expect that the vast majority of dimers will constitute 8 nm GNPs bound to a single DNA molecule that is hybridized to a single 5 nm conjugate (forming dimers with a single dsDNA molecule). Figure 2 shows a gel in which such a separation was carried out. Lanes 1 and 2 are control lanes: lane 1 contains 5 nm GNPs with no DNA bound to them; lane 2 contains 8 nm GNPs with no DNA bound to them. Lane 3 contains the hybridized mixture of the two GNPs/DNA conjugates, which was separated into bands. These bands contain, from bottom up, 5 nm GNPs, 8 nm GNPs, 8 nm+5 nm dimers. The following bands contain higher oligomeric structures, as indicated by the illustrations next to the arrows on the right. In order to visualize and check the formation of dimer structures, the third band from the bottom (framed), which was expected to contain dimers, was extracted from the gel for further analysis by TEM imaging (Figure 3). The majority of the observed structures are of 5–8 nm GNPs dimers. These results demonstrate that the hybridization of additional structures in large excess can assist separation between GNP/DNA conjugates based on the number of short ssDNA strands that they contain.
Figure 1 Effect of DNA length on separation of DNA/GNP conjugates with gel electrophoresis. 8 nm GNPs were attached to thiolated DNA molecules as described in Section 2. (a) Lane 1 contained a control sample of 8 nm GNPs with no DNA. Lane 2 contained conjugates with 86 bases DNA molecules. The conjugate mixtures were separated into bands, each containing a different number of DNA strands as indicated by the illustrations next to the arrows (adapted from [1]). (b) Lane 1 contained a control sample of 8 nm GNPs with no DNA. Lane 2 contains conjugates of 8 nm GNPs with 26 bases DNA molecules (strand S26 1). The conjugate mixtures were not separated into bands in this gel.
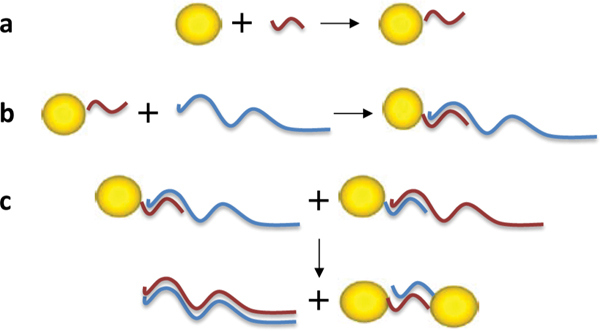
In the second method, we hybridized the 8 nm GNP/26 bases ssDNA (strands S26 1andS26 2) conjugates with long ssDNA molecules (100 bases ssDNA, strands RC100 1 and RC100 2) with a 26 bp overlap, as illustrated in Figure 4. Again, because the long ssDNA molecules were in a large excess one may assume that they hybridized to all the short ssDNA molecules bound to the 8 nm GNPs. Similarly to the previous method, this hybridization enabled conjugates separation based on the number of 26 bases ssDNA molecules that are bound to them, which are now extended by the 100 bases strands.
Figure 2 Electrophoretic separation of 8 nm GNP/26 bases DNA (S26 1) conjugates through binding to 5 nm GNP/26 bases DNA (S26 2) conjugates. Lane 1 contains a control sample of 5 nm GNPs with no DNA. Lane 2 contains a control sample of 8 nm GNPs with no DNA. Lane 3 contains the hybridized mixture of the two types of GNP/DNA conjugates with complementary 26 base DNA strands. The conjugates mixture was separated into bands indicating that the hybridization produced structures of 8 and 5 nm GNPs that are connected by dsDNA as illustrated next to the arrows. The structures in the band in lane 3 marked by the dashed frame were extracted from the gel and imaged by TEM (see Figure 3).
Figure 3 TEM image of 8 and 5 nm GNP dimer structures that are connected by 26 bases dsDNA. The structures were extracted from the gel band marked by the frame in Figure 2. The majority of the structures imaged were dimers of 8 nm GNPs with 5 nm GNPs. Scale bar 20 nm.
Figure 4 Illustration of the procedure for the synthesis of dimers composed of GNPs connected by a single dsDNA molecule. (a) Conjugation: 8 nm GNPs were attached to 26 bases ssDNA. (b) Hybridization with a complementary 100 bases ssDNA over the 26 bp complementary part. Gel electrophoresis was used in order to separate the GNP/DNA conjugates bound to a single ssDNA molecule. (c) The GNP/DNA conjugates were extracted from the gel and mono-conjugates with the complementary short DNA molecules were mixed. By heating and then cooling down the solution, the complementary 100 bases strands form dsDNA and the GNPs that were bound to the short DNA molecule formed dimers. Following hybridization, the structures were again separated by gel electrophoresis (see Figure 5).
Figure 5 shows a gel in which such a separation was carried out. Lane 1 contains 8 nm GNPs with no bound DNA. The other two lanes contain conjugates of 8 nm GNPs with 26 base ssDNA after their hybridization with 100 base DNA molecules. Each one of these lanes contained conjugates with one of the two complementary 26 bases ssDNA strands (strand S26 1 hybridized with RC100 1 and strand S26 2 hybridized with RC100 2, respectively). The conjugate mixtures were separated in this gel into bands, each containing a different number of DNA strands bound to the GNP, as illustrated next to the arrows on the right. This clearly shows that the extension with the longer DNA strands influence the conjugates’ mobility and enables the desired separation. The inset in Figure 5 demonstrates separation of 20 nm GNPs attached to 26 bases ssDNA (strands S26 1) after their hybridization with 100 bases ssDNA strands (strand RC100 1). The right lane (that contains the 100 bases strands) was separated in this gel into bands. Those results demonstrate that the separation method can be affective and useful also for larger GNPs. Comparison of the separation obtained in the lanes that contain 10 nm (the two right lanes in Figure 5) and 20 nm (the right lane in the inset in Figure 5) GNPs shows, however, that the separation efficiency drops for larger GNPs for a given DNA length.
Figure 5 Electrophoretic separation of GNP/26 bases ssDNA hybridized with complementary long DNA molecules. Lane 1 contains a control sample of 8 nm GNPs with no DNA. The other lanes contain conjugates of 8 nm GNPs with each of the two complementary 26 bases ssDNA molecules after their hybridization with 100 base DNA molecules (lane 2 contains 8 nm GNPs attached to S26 1 hybridized to RC100 1, lane 3 contains 8 nm GNPs attached to S26 2 hybridized to RC100 2). The conjugate mixtures were separated in this gel into bands, each containing a different number of DNA strands as indicated by the illustrations next to the arrows to the right. Inset: the left lane contains conjugates of 20 nm GNPs with 26 bases ssDNA molecules (S26 1). The right lane contains conjugates of 20 nm GNPs with 26 bases ssDNA molecules (S26 1) after their hybridization with 100 bases ssDNA molecules (RC100 1).
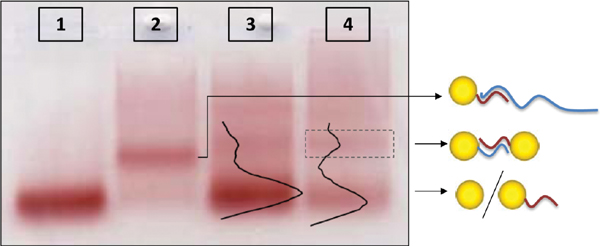
GNP/DNA mono-conjugates were extracted from the second band from the bottom in lanes 2 and 3 in the gel, and mono-conjugates with complementary sequences were mixed. The 100 bases ssDNA strands were de-hybridized from the conjugates by heating, and then the solution was cooled down. During the cooling, the 100 bases ssDNA molecules hybridize to each other at relatively high temperatures, before the temperature is low enough for them to re-hybridize to the short conjugates. Therefore, when the temperature is further decreased the 100 bases strands are not in ssDNA form anymore and not available to rebind to the short 26 bases ssDNA molecules that hybridize to each other to form dimers, as illustrated in Figure 4 and demonstrated in Figure 6. Figure 6 shows the gel in which samples were separated after hybridization of the mono-conjugates. Lanes 1-3 are control lanes. Lane 1 contains a control sample of 8 nm GNPs with no bound DNA; lane 2 contains conjugates of 8 nm GNPs with 26 bp ssDNA (S26 1) hybridized to one 100 bases ssDNA (RC100 1) (similar to the second band in lane 2 in Figure 5) and lane 3 contains a hybridized mixture of 8 nm GNP/26 base ssDNA conjugates, which had not been separated prior to hybridization. Lane 4 contains a hybridized mixture of two complementary mono-conjugates after they were separated by hybridization to 100 bases ssDNA molecules. In lanes 3 and 4 a separation to bands is observed. The bands in these lanes contain from bottom up: mono-conjugates (mixed with bare GNPs), dimers and higher oligomeric structures. In lane 4 there is no band parallel to the one in lane 2 (monoconjugates of 8 nm GNPs bound to 26 bp DNA strand that hybridized to 100 bases complementary DNA). This indicates clearly that the hybridization between two 100 bases DNA strands can effectively release the 26 bases ssDNA bound to the GNP so they are free to form dimers. Note that the total “color” in lane 3 that did not pass a first separation is stronger, as it overall contains more GNPs, but the relative intensity of the dimer band (second from bottom) in lane 4 compared to the bare GNP band (first band) is stronger than in lane 3 since after the first separation the relative amount of dimers is larger.
Figure 6 Separation of DNA/GNP conjugates by gel electrophoresis. Lanes 1–3 contain control samples: Lane 1 contains a sample of 8 nm GNPs with no bound DNA. Lane 2 contains conjugates of 8 nm GNPs with 26 bp DNA sequences (S26 1) hybridized to 100 bases ssDNA (RC100 1); lane 3 contains a hybridized mixture of 8 nm GNPs that are bound to 26 bases complementary ssDNA molecules (8 nm GNPs attached to S26 1 hybridized to 8 nm GNPs attached to S26 2). In this case, the conjugates might contain varying numbers of DNA molecules. Lane 4 contains a hybridized mixture of two complementary mono-conjugates (8 nm GNPs attached to single S26 1 and to 8 nm GNPs attached to single S26 2) after they were separated by hybridization to long DNA (RC100 1 and RC 100 2 respectively) so it is verified that only one dsDNA connects the GNPs in each dimer. The arrows point to illustrations of the expected structures.
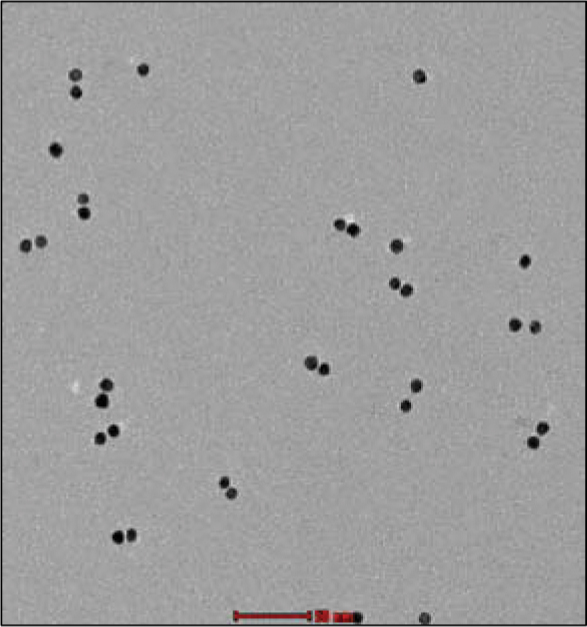
Figure 7 TEM image of the synthesized GNP dimers after using the separation method described in the text. The structures were extracted from the gel band indicated by the frame in Figure 6, lane 4. Most of the particles are assembled into dimers. Scale bar 50 nm.
The bands that were expected to contain dimers were extracted from the gel for further analysis by TEM. TEM characterization of the dimer band in lane 4 is presented in Figure 7. The TEM imaging demonstrates that dimers were indeed formed and that the majority of the particles belong to dimers. Though the second band from the bottom in both lanes 3 and 4 was shown (by TEM characterization) to contain dimer structures, one must note that while the GNPs forming the dimers in lane 3 may be attached by more than one dsDNA molecule and may have additional ssDNA molecules bound to them, the dimers in lane 4 have only one dsDNA molecule attaching the GNPs and no additional ssDNA molecules. This property cannot be characterized either by gel electrophoresis or by TEM, but it is essential and stems from our synthesis procedure.
4. Conclusions
When GNPs and DNA are mixed in 1:1 ratio, at least some of the GNPs are connected to more than one DNA strand. Therefore, dimers directly formed by mixing these conjugates may be connected by more than a single dsDNA, and may have additional ssDNA molecules bound to them. When the DNA molecule is long, gel electrophoresis can be used for separation between GNPs attached to different numbers of DNA strands. When short DNA strands are connected to the GNPs, however, the DNA/GNPs monoconjugates cannot be directly separated by gel electrophoresis. We show here two simple methods by which one can efficiently separate conjugates bearing different numbers of short DNA strands. These two methods provide the requested resolution in gel electrophoresis and enable the isolation of mono-conjugates containing short DNA strands.
Each one of the two presented methods has its own advantages. In the first method the DNA/GNP conjugates were hybridized to complementary ssDNA molecules connected to smaller GNPs. This method allows validation of the process at intermediate stages. After the separation stage, for example, the dimers (formed from monoconjugates hybridized to only one complementary ssDNA attached to a small GNP) can be imaged by TEM. In the second method the DNA/GNP conjugates were hybridized with long complementary ssDNA. This method enables easy and convenient release of the extending strand from the conjugates for further use of the mono-conjugates. In our work, for example, they were used for the formation of dimers composed of two equal sized GNPs connected by a single dsDNA.
The two methods we demonstrate here for separation of 8 nm GNPs conjugated to 26 base ssDNA can be implemented also for larger GNPs. As the GNP diameter increases the separation efficiency for a given DNA length drops (as demonstrated in the inset in Figure 5). Effective separation can be maintained, however, by simply increasing the length of attached DNA by hybridization with longer complementary DNA strands. In addition to dimers formation, nanoparticles with a discrete number of DNA molecules can be used as building blocks for a variety of nanostructures. The methods we present here may be implemented for preparation of such nanostructures.
Acknowledgments
This work was supported by the European Commission through FP6-IST grant for Future & Emerging Technologies ‘DNA-based Nanodevices’ (FP6-029192); the ESF COST MP0802; the Israel Science Foundation (grant no. 1145/10); BSF grant 2006422; the French Ministry of External Affairs, the Israeli-Palestinian Science Organization and Friends of IPSO, USA (with funds donated by the Meyer Foundation); Academia Sinica and the Hebrew University of Jerusalem joint research program in Nanoscience and Nanotechnology. HD thanks the Klein foundation for MSc scholarships.
References
[1] A. Stern, D. Rotem, I. Popov, D. Porath Journal of Physics: Condensed Matter, 24(16), 164203 (2012).
[2] C. J. Loweth, W. B. Caldwell, X. Peng, A. P. Alivisatos, P. G. Schultz Angewandte Chemie International Edition, 38(12), 1808–1812 (1999).
[3] J. Zheng, P. E. Constantinou, C. Micheel, A. P. Alivisatos, R. A. Kiehl, N. C. Seeman Nano Letters, 6(7), 1502–1504 (2006).
[4] H. Cohen, C. Nogues, R. Naaman, D. Porath Proceedings of the National Academy of Sciences of the United States of America, 102(33), 11589–11593 (2005).
[5] H. Cohen, C. Nogues, D. Ullien, S. Daube, R. Naaman, D. Porath Faraday Discuss., 131, 367–376 (2005).
[6] D. Ullien, H. Cohen, D. Porath Nanotechnology, 18(42), 424015 (2007).
[7] A. Kuzyk, R. Schreiber, Z. Fan, G. Pardatscher, E. M. Roller, A. Högele, F. C. Simmel, A. O. Govorov, T. Liedl Nature, 483(7389), 311–314 (2012).
[8] J.Wirth,F.Garwe,G.Hähnel, A. Csaki, N. Jahr, O. Stranik, W. Paa, W. Fritzsche Nano Letters, 11(4), 1505–1511 (2011).
[9] T. Song, H. Liang Journal of the American Chemical Society, 134(26), 10803–10806 (2012).
[10] J. J. Storhoff, A. A. Lazarides, R. C. Mucic, C. A. Mirkin, R. L. Letsinger, G. C. Schatz Journal of the American Chemical Society, 122(19), 4640–4650 (2000).
[11] J. Liu, Y. Lu Journal of the American Chemical Society, 125(22), 6642–6643 (2003).
[12] W. Zhao, C. Yao, X. Luo, L. Lin, I. Hsing Electrophoresis, 33(8), 1288–1291 (2012).
[13] D. A. Giljohann, D. S. Seferos, W. L. Daniel, M. D. Massich, P. C. Patel, C. A. Mirkin Angewandte Chemie International Edition, 49(19), 3280–3294 (2010).
[14] J. Xue, L. Shan, H. Chen, Y. Li, H. Zhu, D. Deng, Z. Qian, S. Achilefu, Y. Gu Biosensors and Bioelectronics (2012).
[15] C. L. Choi, A. P. Alivisatos Annual Review of Physical Chemistry, 61, 369–389 (2010).
[16] D. Zanchet, C. M. Micheel, W. J. Parak, D. Gerion, A. P. Alivisatos Nano Letters, 1(1),32–35 (2001).
[17] W. Zhao, I. M. Hsing Chemical Communications, 46(8), 1314–1316 (2010).
[18] N. Borovok, E. Gillon, A. Kotlyar Bioconjugate Chemistry, 23(5), 916–922 (2012).
[19] M. P. Busson, B. Rolly, B. Stout, N. Bonod, E. Larquet, A. Polman, S. Bidault Nano Letters, 11(11), 5060–5065 (2011).
[20] S. A. Claridge, H. W. Liang, S. R. Basu, J. M. J. Fréchet, A. P. Alivisatos Nano Letters, 8(4), 1202–1206 (2008).
[21] A. J. Mastroianni, S. A. Claridge, A. P. Alivisatos Journal of the American Chemical Society, 131(24), 8455–8459 (2009).
Biographies

Haya Dachlika got her B.Sc degree in Chemistry and Biology from The Hebrew University of Jerusalem, Israel in 2010. In 2011 Haya started her master’s degree in Chemistry at The Hebrew University of Jerusalem under supervision of Professor Danny Porath. Her research is focused on novel DNA-based functionalized metal nanoparticles.

Avigail Stern got her B.Sc degree in Biology and Chemistry from the Hebrew University of Jerusalem, Israel, in 2010. Avigail is pursuing her Ph.D. in Chemistry at the Hebrew University of Jerusalem under the supervision of Professor Danny Porath. Avigail’s research is focused on charge transport measurements through DNA molecules.

Dvir Rotem got his Ph.D. in Brain and Behavioral Sciences from the Hebrew University of Jerusalem. He joined, as a post-doc, the Bayley group at the Chemistry Department at The University of Oxford, UK in 2005. Since 2011 he is a research associate in the Institute of Chemistry in the Hebrew University of Jerusalem and works as the laboratory manager of Professor Danny Porath’s group.

Danny Porath got his Ph.D. in Physics at the Hebrew University of Jerusalem in 1997. Pioneered electrical transport measurements in single DNA molecules as a postdoc in the group of Professor Cees Dekker at Delft University of Technology. A faculty member at the Hebrew University of Jerusalem since 2001. His major research interest is implementation of bio-molecules in molecular electronics devices in general and understanding the electrical charge transport mechanisms in DNA in particular.