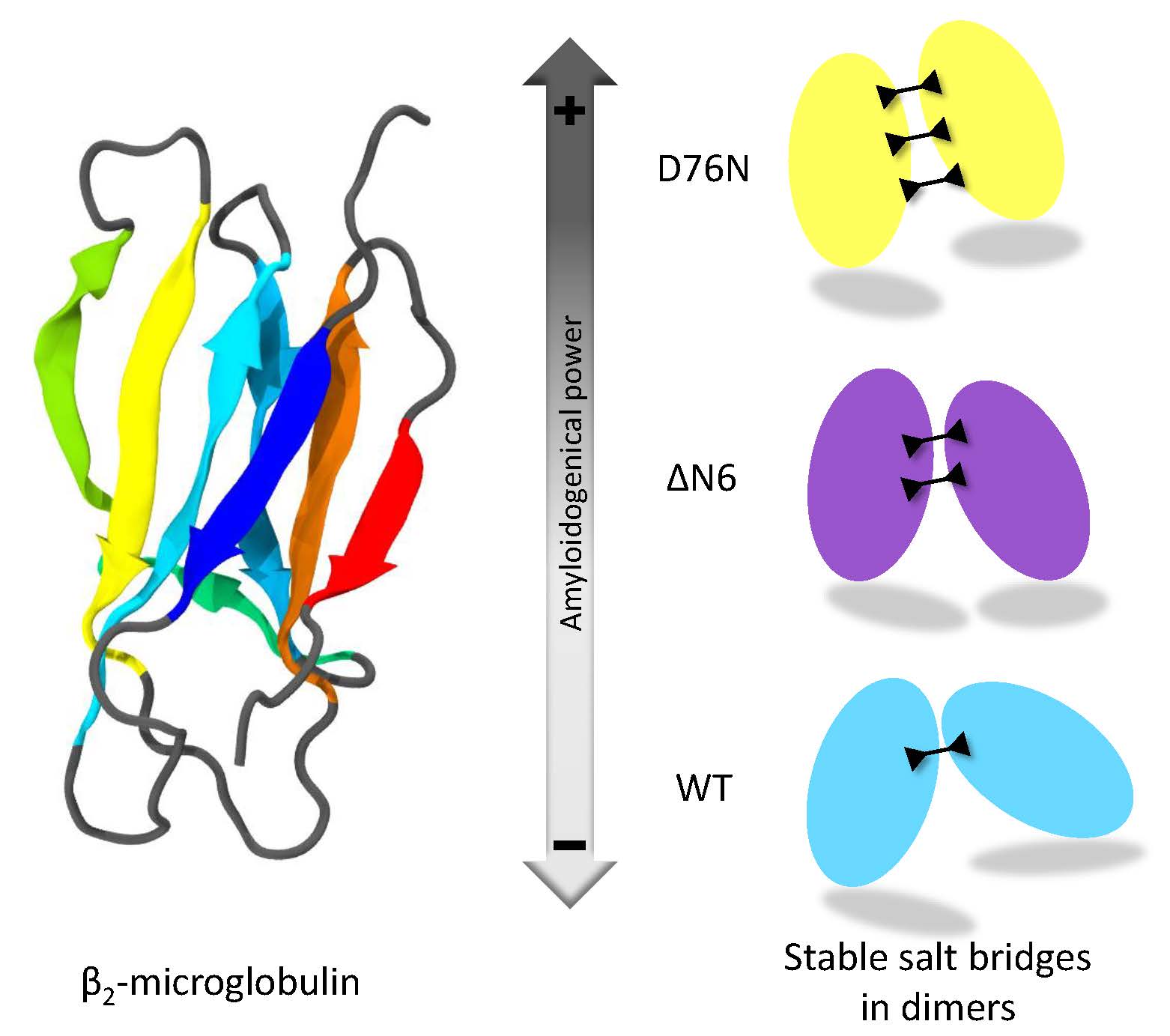
Salt Bridges Regulate in Silico Dimers Formation for β2-Microglobulin Amyloidogenic Variants
DOI:
https://doi.org/10.13052/jsame2245-4551.6.003Keywords:
amyloidosis, molecular dynamics, protein aggregation, β2-microglobulin.Abstract
β2-microglobulin is a paradigmatic amyloidogenic protein responsible for
dialysis-related amyloidosis, a disease associated to long-term hemodialyzed
patients and characterized by accumulation of amyloid deposits in the osteoar-
ticular tissues. In the early stages of amyloid fibril formation, β2-microglobulin
associates into dimers and higher oligomers, but clarifications are still needed
for the triggering conditions, mechanisms and specificity of dimer forma-
tion. To characterize the dimeric association process, the protein-protein
interactions between three different species are investigated: namely, the
native protein and the two amyloidogenic variants ΔN6 and D76N. The
dimerization process is rationalized relying on state of the art computational
methods. A comparative mechanism for how different mutations in the three
variants can affect protein dimerization and thus fibril formation is proposed. The number of salt bridges involved at the protein-protein interface correlates
with the degree of amyloidogenicity of each individual species. The findings
can offer possible strategies in controlling the dimerization mechanism based
on different β2-microglobulin protein mutations, which have significant roles
in the fibrillogenical process.
Downloads
References
P. J. Bjorkman, M. A. Saper, B. Samraoui, W. S. Bennett, J. L.
Strominger, and D. C. Wiley. Structure of the human class I histocom-
patibility antigen, HLA-A2. Nature, 329:506, 1987.
L. Halabelian, S. Ricagno, S. Giorgetti, C. Santambrogio, A. Barbiroli,
S. Pellegrino, A. Achour, R. Grandori, L. Marchese, S. Raimondi,
P. P. Mangione, G. Esposito, R. Al-Shawi, J. P. Simons, I. Speck,
M. Stoppini, M. Bolognesi, and V. Bellotti. Class I major histocom-
patibility complex, the trojan horse for secretion of amyloidogenic
β2-microglobulin. Journal of Biological Chemistry, 289(6):3318–3327,
F. Gejyo, T. Yamada, S. Odani, Y. Nakagawa, M.Arakawa, T. Kunitomo,
H. Kataoka, M. Suzuki, Y. Hirasawa, T. Shirahama, A. S. Cohen, and
K. Schmid. A new form of amyloid protein associated with chronic
hemodialysis was identified as β2-microglobulin. Biochemical and
Biophysical Research Communications, 129(3):701–706, 1985.
A. Corazza, E. Rennella, P. Schanda, M. C. Mimmi, T. Cutuil,
S. Raimondi, S. Giorgetti, F. Fogolari, P. Viglino, L. Frydman, M. Gal,
V. Bellotti, B. Brutscher, and G. Esposito. Native-unlike long-lived
intermediates along the folding pathway of the amyloidogenic pro-
tein β2-microglobulin revealed by real-time two-dimensional NMR.
Journal of Biological Chemistry, 285(8):5827–5835, 2010.
C. Rosano, S. Zuccotti, and M. Bolognesi. The three-dimensional
structure of β2-microglobulin: results from x-ray crystallography.
Biochimica et Biophysica Acta, 1753(1):85–91, 2005.
M. Stoppini and V. Bellotti. Systemic amyloidosis: Lessons from
β2-microglobulin. Journal of Biological Chemistry, 290(16):9951–
, 2015.
H. Naiki, N. Hashimoto, S. Suzuki, H. Kimura, K. Nakakuki, and
F. Gejyo. Establishment of a kinetic model of dialysis-related amyloid
fibril extension in vitro. Amyloid, 4(4):223–232, 1997.
M. C. Maschio et al.
C. M. Eakin, A. J. Berman, and A. D. Miranker. A native to amy-
loidogenic transition regulated by a backbone trigger. Nature Structural
Molecular Biology, 13:202, 2006.
M. Stoppini, P. Mangione, M. Monti, S. Giorgetti, L. Marchese,
P. Arcidiaco, L. Verga, S. Segagni, P. Pucci, G. Merlini, and V. Bellotti.
Proteomics of β2-microglobulin amyloid fibrils. Biochimica et Biophys-
ica Acta, 1753(1):23–33, 2005.
G. Esposito, R. Michelutti, G. Verdone, P. Viglino, H. Hern ́andez,
C.V. Robinson, A. Amoresano, F. Dal Piaz, M. Monti, P. Pucci,
P. Mangione, M. Stoppini, G. Merlini, G. Ferri, and V. Bellotti. Removal
of the n-terminal hexapeptide from human β2-microglobulin facilities
protein aggregation and fibril formation. Protein Science, 9(5):831–845,
V. Bellotti, M. Gallieni, S. Giorgetti, and D. Brancaccio. Dynamic of β2-
microglobulin fibril formation and reabsorption: The role of proteolysis.
Seminars in Dialysis, 14(2):117–122, 2001.
T. Eichner, A. P. Kalverda, G. S. Thompson, S. W. Homans, and
S. E. Radford. Conformational conversion during amyloid formation
at atomic resolution. Molecular Cell, 41(2):161–172, 2011.
S. Valleix, J. D. Gillmore, F. Bridoux, P. P . Mangione, A. Dogan,
B. Nedelec, M. Boimard, G. Touchard, J. M. Goujon, C. Lacombe,
P. Lozeron, D.Adams, C. Lacroix, T. Maisonobe, V. Plante-Bordeneuve,
J. A. Vrana, J. D. Theis, S. Giorgetti, R. Porcari, S. Ricagno,
M. Bolognesi, M. Stoppini, M. Delpech, M. B. Pepys, P. N. Hawkins,
and V. Bellotti. Hereditary systemic amyloidosis due to Asp76Asn
variant β2-microglobulin. New England Journal of Medicine, 366(24):
–2283, 2012.
M. de Rosa,A. Barbiroli, S. Giorgetti, P. P. Mangione, M. Bolognesi, and
S. Ricagno. Decoding the structural bases of D76N β2-microglobulin
high amyloidogenicity through crystallography and ASN-scan mutage-
nesis. PLosOne, 1–15, 2015.
P. P. Mangione, G. Esposito, A. Relini, S. Raimondi, R. Porcari,
S. Giorgetti, A. Corazza, F. Fogolari, A. Penco, Y. Goto, Y-H. Lee,
H. Yagi, C. Cecconi, M. M. Naqvi, J. D. Gillmore, P. N. Hawkins,
F. Chiti, R. Rolandi, G. W. Taylor, M. B. Pepys, M. Stoppini, and
V. Bellotti. Structure, folding dynamics, and amyloidogenesis of D76N
β2-microglobulin: roles of shear flow, hydrophobic surfaces and
α-crystallin. Journal of Biological Chemistry, 288(43):30917–30930,
Salt Bridges Regulate in Silico Dimers Formation 55
K. Domanska, S. Vanderhaegen, V. Srinivasan, E. Pardon, F. Dupeux,
J. A. Marquez, S. Giorgetti, M. Stoppini, L. Wyns, V. Bellotti, and
J. Steyaert. Atomic structure of a nanobody-trapped domain-swapped
dimer of an amyloidogenic β2-microglobulin variant. Proceedings of
the National Academy of Sciences, 108(4):1314–1319, 2011.
D. P. Smith, S. Jones, L. C. Serpell, M. Sunde, and S. E. Radford.
A systematic investigation into the effect of protein destabilisation on
β2-microglobulin amyloid formation. Journal of Molecular Biology,
(5):943–954, 2003.
T. R. Jahn, M. J. Parker, S. W. Homans, and S. E. Radford. Amyloid
formation under physiological conditions proceeds via a native-like
folding intermediate. Nature Structural and Molecular Biology, 13,
T. Eichner and S. E. Radford. A generic mechanism of β2-microglobulin
amyloid assembly at neutral pH involving a specific proline switch.
Journal of Molecular Biology, 386(5):1312–1326, 2009.
G. W. Platt and S. E. Radford. Glimpses of the molecular mechanisms
of β2-microglobulin fibril formation in vitro: aggregation on a complex
energy landscape. FEBS Letter, 583(16):2623–2629, 2009.
N. H. H. Heegaard, T. J. D. Jørgensen, N. Rozlosnik, D. B. Corlin,
J. S. Pedersen, A. G. Tempesta, P. Roepstorff, R. Bauer, and M. H. Nis-
sen. Unfolding, aggregation, and seeded amyloid formation of Lysine-
-cleaved β2-microglobulin. Biochemistry, 44(11):4397–4407, 2005.
S. Giorgetti, S. Raimondi, K. Pagano, A. Relini, M. Bucciantini,
A. Corazza, F. Fogolari, L. Codutti, M. Salmona, P. Mangione,
L. Colombo,A. De Luigi, R. Porcari,A. Gliozzi, M. Stefani, G. Esposito,
V. Bellotti, and M. Stoppini. Effect of tetracyclines on the dynamics
of formation and destructuration of β2-microglobulin amyloid fibrils.
Journal of Biological Chemistry, 286(3):2121–2131, 2011.
F. Fogolari, A. Corazza, P. Viglino, P. Zuccato, L. Pieri, P. Faccioli,
V. Bellotti, and G. Esposito. Molecular dynamics simulation sug-
gests possible interaction patterns at early steps of β2-microglobulin
aggregation. Biophysical Journal, 92(5):1673–1681, 2007.
S. G. Estacio, H. Krobath, D. Vila-Vicosa, M. Machuqueiro,
E. I. Shakhnovich, and P. F. N. Faisca. A simulated intermediate state for
folding and aggregation provides insights into ΔN6 β2-microglobulin
amyloidogenic behavior. PLoS Computational Biology, 10(5):1–17,
M. C. Maschio et al.
C. M. Eakin, F. J. Attenello, C. J. Morgan, and A. D. Miranker.
Oligomeric assembly of native-like precursors precedes amyloid for-
mation by β2-microglobulin. Biochemistry, 43(24):7808–7815, 2004.
A. M. Smith, T. R. Jahn, A. E. Ashcroft, and S. E. Radford. Direct obser-
vation of oligomeric species formed in the early stages of amyloid fibril
formation using electrospray ionisation mass spectrometry. Journal of
Molecular Biology, 364(1):9–19, 2006.
H. E. White, J. L. Hodgkinson, T. R. Jahn, S. Cohen-Krausz,
W. S. Gosal, S. M ̈uller, E. V. Orlova, S. E. Radford, and H. R.
Saibil. Globular tetramers of β2-microglobulin assemble into elaborate
amyloid fibrils. Journal of Molecular Biology, 389(1):48–57, 2009.
E. Rennella, T. Cutuil, P. Schanda, I. Ayala, F. Gabel, V. Forge,
A. Corazza, G. Esposito, and B. Brutscher. Oligomeric states along the
folding pathways of β2-microglobulin: kinetics, thermodynamics, and
structure. Journal of Molecular Biology, 425(15):2722–2736, 2013.
M. Martinez, N. J. Bruce, J. Romanowska, D. B. Kokh, M. Ozboyaci,
X. Yu, M. A. ̈Ozt ̈urk, S. Richter, and R. C. Wade. SDA 7: a modular
and parallel implementation of the simulation of diffusional association
software. Journal of Computational Chemistry, 36(21):1631–1645,
D. J. Earl and M. W. Deem. Parallel tempering: Theory, applications and
new perspectives. Physical Chemistry Chemical Physics, 7:3910–3916,
G. Verdone, A. Corazza, P. Viglino, F. Pettirossi, S. Giorgetti,
P. P. Mangione, A. Andreola, M. Stoppini, V. Bellotti, and G. Espos-
ito. The solution structure of human β2-microglobulin reveals the
prodromes of its amyloid transition. Protein Science, 11(3):487–499,
D. Gumral, F. Fogolari,A. Corazza, P. Viglino, S. Giorgetti, M. Stoppini,
V. Bellotti, and G. Esposito. Reduction of conformational mobility
and aggregation in W60G β2-microglobulin: assessment by N15 NMR
relaxation. Magnetic Resonance in Chemistry, 51(12):795–807, 2015.
G. Esposito, M. Garvey, V.Alverdi, F. Pettirossi,A. Corazza, F. Fogolari,
M. Polano, P. P. Mangione, S. Giorgetti, M. Stoppini, A. Rekas,
V. Bellotti, A.J.R. Heck and J. A. Carver. Monitoring the interac-
tion between β2-microglobulin and the molecular chaperone αB-
crystallin by NMR and mass spectrometry: αb-crystallin dissociates
β2-microglobulin oligomers. The Journal of Biological Chemistry,
(24):17844–17858, 2013.
Salt Bridges Regulate in Silico Dimers Formation 57
G. Brancolini, D. Toroz, and S. Corni. Can small hydrophobic
gold nanoparticles inhibit β2-microglobulin fibrillation? Nanoscale,
: 7903–7911, 2014.
L. Halabelian, A. Relini, A. Barbiroli, A. Penco, M. Bolognesi, and S.
Ricagno. A covalent homodimer probing early oligomers along amyloid
aggregation. Scientific Reports, 5, 2015.
M. Beerbaum, M. Ballaschk, N. Erdmann, C. Schnick, A. Diehl,
B. Uchanska-Ziegler, A. Ziegler, and P. Schmieder. NMR spectroscopy
reveals unexpected structural variation at the protein-protein interface in
MHC class I molecules. Journal of Biomolecular NMR, 57(2):167–178,
R. J. S. Loureiro, D. Vila-Vicosa, M. Machuqueiro, E. I. Shakhnovich,
and P. F. N. Faisca. A tale of two tails: The importance of unstructured
termini in the aggregation pathway of β2-microglobulin. Proteins:
Structure, Function, and Bioinformatics, 85(11):2045–2057, 2017.
S. Raimondi, R. Porcari, P. P. Mangione, G. Verona, J. Marcoux,
S. Giorgetti, G. W. Taylor, S. Ellmerich, M. Ballico, S. Zanini, E. Pardon,
R. Al-Shawi, J. P. Simons, A. Corazza, F. Fogolari, M. Leri, M. Stefani,
M. Bucciantini, J. D. Gillmore, P. N. Hawkins, M. Valli, M. Stoppini,
C. V. Robinson, J. Steyaert, G. Esposito, and V. Bellotti. A specific
nanobody prevents amyloidogenesis of D76N β2-microglobulin in vitro
and modifies its tissue distribution in vivo. Scientific Reports, 7:46711,
A specific nanobody prevents amyloidogenesis of D76N The control-
ling roles of Trp60 and Trp95 in β2-microglobulin function, folding
and amyloid aggregation properties. Journal of Molecular Biology,
:887, 2008.
V. L. Mendoza, K. Antwi, M. A. Bar ́on-Rodr ́iguez, C. Blanco, and
R. W. Vachet. Structure of the preamyloid dimer of β2-microglobulin
from covalent labeling and mass spectrometry. Biochemistry, 49(7):
–1532, 2010.
G. W. Platt, K. E. Routledge, S. W. Homans, and S. E. Radford.
Fibril growth kinetics reveal a region of β2-microglobulin important
for nucleation and elongation of aggregation. Journal of Molecular
Biology, 378(1):251–263, 2008.
T. Eichner and S. E. Radford. Understanding the complex mecha-
nisms of β2-microglobulin amyloid assembly. FEBS Journal, 278(20):
–3883, 2011.
M. C. Maschio et al.
A. H. Elcock, R. R. Gabdoulline, R. C. Wade, and J. A. McCammon.
Computer simulation of protein-protein association kinetics:
acetylcholinesterase-fasciculin. Journal of Molecular Biology,
(1):149–162, 1999.
D. Van Der Spoel, E. Lindahl, B. Hess, G. Groenhof, A. E. Mark,
and H. J. C. Berendsen. Gromacs: Fast, flexible, and free. Journal of
Computational Chemistry, 26(16):1701–1718, 2005.
G. Bussi, D. Donadio, and M. Parrinello. Canonical sampling through
velocity rescaling. Journal of Chemical Physics, 126(1), 2007.
M. Parrinello and A. Rahman. Polymorphic transitions in single crys-
tals: A new molecular dynamics method. Journal of Applied Physics,
(12):7182–7190, 1981.
G. Brancolini, D. B. Kokh, L. Calzolai, R. C. Wade, and S. Corni. Dock-
ing of ubiquitin to gold nanoparticles. ACS Nano, 6(11):9863–9878,
G. Brancolini, A. Corazza, M. Vuano, F. Fogolari, M. C. Mimmi,
V. Bellotti, M. Stoppini, S. Corni, and G. Esposito. Probing the influence
of citrate-capped gold nanoparticles on an amyloidogenic protein. ACS
Nano, 9(3):2600–2613, 2015.
C. Cantarutti, S. Raimondi, G. Brancolini, A. Corazza, S. Giorgetti,
M. Ballico, S. Zanini, G. Palmisano, P. Bertoncin, L. Marchese, P. P.
Mangione, V. Bellotti, S. Corni, F. Fogolari, and G. Esposito. Citrate-
stabilized gold nanoparticles hinder fibrillogenesis of a pathological
variant of β2-microglobulin. Nanoscale, 9:3941–3951, 2017.
G. Brancolini, M. C. Maschio, C. Cantarutti, A. Corazza, F. Fogolari,
V. Bellotti, S. Corni, and G. Esposito. Citrate stabilized gold
nanoparticles interfere with amyloid fibril formation: D76N and δN6
β2-microglobulin variants. Nanoscale, 10:4793–4806, 2018.
A. Patriksson and D. van der Spoel. A temperature predictor for
parallel tempering simulations. Physical Chemistry Chemical Physics,
:2073–2077, 2008.
M. M. Seibert, A. Patriksson, B. Hess, and D. van der Spoel. Repro-
ducible polypeptide folding and structure prediction using molecular
dynamics simulations. Journal of Molecular Biology, 354(1):173–183,
D. van der Spoel and M. M. Seibert. Protein folding kinetics and
thermodynamics from atomistic simulations. Physical Review Letter.,
:238102, 2006.
S. Kirkpatrick, C. D. Gelatt, and M. P. Vecchi. Optimization by
simulated annealing. Science, 220(4598):671–680, 1983