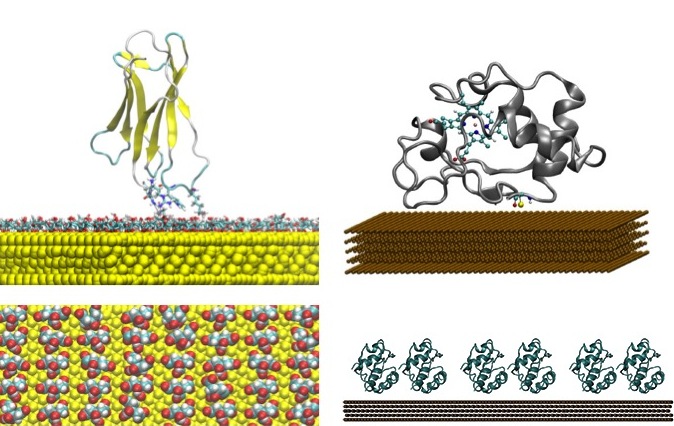
Computational Strategies for Protein-Surface and Protein-Nanoparticle Interactions
DOI:
https://doi.org/10.13052/jsame2245-4551.211Keywords:
gold; nanoparticles; amyloid; docking, molecular dynamics, redox potential.Abstract
Protein-nanoparticle associations have important applications in nanoscience
and nanotechnology but the recognition mechanisms and the determinants
of specificity are still poorly understood at the microscopic level. Crucial
questions remain open, related to the association mechanisms, control of
binding events, and preservation of functionality. Gold is a promising material
in nanoparticles for nanobiotechnology applications because of the ease of
its functionalization and its tunable optical properties. We present a concise
overview of recent computational modeling advances which were pursued in
the quest for a theoretical framework elucidating the association mechanisms
and the ability to design and control the recognition events of a specific
class of systems, namely, interfaces between polypeptides/proteins and a gold
surface in the presence of water. We select two different methodological
advances, the first related to the effect of surfactants covering the surface
of nanoparticles and altering their interactions with proteins and the second
related to the immobilization of proteins on inorganic surfaces and conserving
their functionality. Both cases, demonstrate how the understanding of the
polypeptide-surface coupling mechanisms is essential to the control of the
process and exploitation for biotechnological and nanotechnological purposes.
Downloads
References
M. Sarikaya, C. Tamerler, A. K. Jen, K. Schulten, F. Baneyx, Molec-
ular Biomimetics: Nanotechnology through Biology. Nat. Mater., 2,
–585 (2003).
S. Linse, C. Cabaleiro-Lago, W.-F. Xue, I. Lynch, S. Lindman,
E. Thulin, S. E. Radford, K. A. Dawson, Nucleation of protein fibrillation
by nanoparticles. Proc. Natl. Acad. Sci. USA, 104, 8691–8696 (2007).
M. Mahmoudi, I. Lynch, M. R. Ejtehadi, M. P. Monopoli, F. B.
Bombelli, S. Laurent, Protein-Nanoparticle Interactions: Opportunities
and Challenges. Chem. Rev., 111, 5610–5637 (2011).
M.-E. Aubin-Tam, S. Hwang, and K Hamad-Schifferli, Site-Directed
Nanoparticle labeling of Cytochrome C. Proc. Natl. Acad. Sci. U.S.A.,
, 4095–4100 (2009).
G. Goobes, R. Goobes, O. Schueler-Furman, D. Baker, P. S. Stayton,
Drobny, G. P. Folding of the C-Terminal Bacterial binding Domain in
Statherin upon Adsorption onto Hydroxyapatite Crystals. Proc. Natl.
Acad. Sci. U.S.A., 103, 16083–16088 (2006).
J. J. Gray, The Interaction of Proteins with Solid Surfaces. Curr. Opin.
Struct. Bio., 14, 110–115 (2004).
J. H. Harding, D. M. Duffy, Sushko, L. Maria, P. Mark
Rodger, D. Quigley, J. A. Elliott, Computational techniques at the
organic-inorganic interface in biomineralization. Chem. Rev., 108,
–4854 (2008).
Computational Strategies for Protein-Surface 21
R. A. Latour, Molecular Simulation of Protein-Surface Interactions:
Benefits, Problems, Solutions, and Future Directions. Biointerphases,
, FC2-FC12 (2008).
O. Cohavi, S. Corni, F. De Rienzo, R. Di Felice, K. E. Gottschalk,
M. Hoefling, D. Kokh, E. Molinari, G. Schreiber, A. Vaskevich, R. C.
Wade, Protein-Surface Interactions: Challenging Experiments and Com-
putations. J. Mol. Recognit., 23, 259–262 (2009).
M. Bachmann, K. Goede, A. G. Beck-Sickinger, M. Grundmann,
A. Irb ̈ack, and W. Janke, Microscopic Mechanism of Specific Peptide
Adhesion to Semiconductor Substrates. Angew. Chem., Int. Ed., 49,
–9533 (2010).
R. Di Felice, S. Corni, Simulation of Peptide-Surface Recognition.
J. Phys. Chem. Lett., 2, 1510–1519 (2011).
K. Makrodimitris, D. L. Masica, E. T. Kim, J. J. Gray, Structure
Prediction of Protein-Solid Surface Interactions Reveals a Molecular
Recognition Motif of Statherin for Hydroxyapatite. J. Am. Chem. Soc.,
, 13713–13722 (2007).
L. M. Ghiringhelli, B. Hess, N. F. A. van der Vegt, L. Delle Site,
Competing Adsorption between Hydrated Peptides and Water onto Metal
Surfaces: from Electronic to Conformational Properties. J. Am. Chem.
Soc., 130, 13460–13464 (2008).
G. Hong, H. Heinz, R. R. R. Naik, B. L. Farmer, R. Pachter,
Toward Understanding Amino Acid Adsorption at Metallic Interfaces:
a Density Functional Theory Study. ACS Appl. Mater. Interfaces, 1,
–392 (2009).
A. Vila Verde, J. M. Acres, J. K. Maranas, Investigating the Specificity
of Peptide Adsorption on Gold Using Molecular Dynamics Simulations.
Biomacromolecules, 10, 2118–2128 (2009).
A. Vila Verde, P. J. Beltramo, J. K. Maranas, Adsorption of
Homopolypeptides on Gold Investigated Using Atomistic Molecular
Dynamics. Langmuir, 27, 10, 5918–5926 (2011).
R. Coppage, J. M. Slocik, B. D. Briggs, A. I. Frenkel, H. Heinz,
R. R. Naik, M. R. Knecht, Crystallographic Recognition Controls Pep-
tide Binding for Bio-Based Nanomaterials. J. Am. Chem. Soc., 133,
–12349 (2011).
A. Calzolari, G. Cicero, C. Cavazzoni, R. D. Felice, A. Catellani,
S. Corni, Hydroxyl-Rich β-Sheet Adhesion to the Gold Surface
in Water by First-Principle Simulations. J. Am. Chem. Soc., 132,
–4795 (2010).
J. Yu, M. L. Becker, G. Carri, The Influence of Amino Acid Sequence and
Functionality on the Binding Process of Peptides onto Gold Surfaces.
Langmuir, 28, 1408–1417 (2012).
G. Brancolini et al.
L. Ruan, H. Ramezani-Dakhel, C. Y. Chiu, E. Zhu, Y. Li, H. H. Y.
Heinz, Tailoring molecular specificity toward a crystal facet: a lesson
from biorecognition toward Pt(111). Nano Lett., 13, 840–846 (2013).
M. Hoefling, F. Iori, S. Corni, K. E. Gottschalk, The Conforma-
tions of Amino Acids on a Gold(111) Surface. ChemPhysChem, 11,
–1767 (2010).
M. Hoefling, Iori F.; S. Corni, K. E. Gottschalk, Interaction of Amino
Acids with the Au(111) Surface: Adsorption Free Energies from Molec-
ular Dynamics Simulations. Langmuir, 26, 8347–8351 (2010).
M. Hoefling, S. Monti, S. Corni, K. E. Gottschalk, Interaction of
β-Sheet Folds with a Gold Surface. PLoS One, 6, e20925 (2012).
D. Toroz, S. Corni, Peptide Synthesis of Gold Nanoparticles: the Early
Steps of Gold Reduction Investigated by Density Functional Theory.
Nano Lett., 11, 1313–1318 (2011).
L. B. Wright, P. M. Rodger, S. Corni, T. R. Walsh, GolP-CHARMM:
First-Principles Based Force Fields for the Interaction of Proteins with
Au(111) and Au(100) J. Chem. Theory Comput., 9, 1616–1630 (2013).
L. B. Wright, P. M. Rodger, T. R. Walsh, S. Corni, First-
Principles-Based Force Field for the Interaction of Proteins with
Au(100)(5x1): An Extension of GolP-CHARMM J. Phys. Chem. C, 117,
–24306 (2013).
G. Brancolini, D. B. Kokh, L. Calzolai, R. C. Wade, S. Corni, Docking
of Ubiquitin to Gold Nanoparticles. ACS NANO, 6, 9863–9878 (2012).
G. Brancolini, D. Toroz, S. Corni, Can small hydrophobic gold nanopar-
ticles inhibit β2-Microglobulin fibrillation? Nanoscale, 6, 7903–7911
(2014).
F. Iori, R. Di Felice, E. Molinari, S. Corni, GolP: An Atomistic Force-
Field to Describe the Interaction of Proteins with Au(111) Surfaces in
Water J. Comp. Chem., 30, 1465–1476 (2009).
M. Aschi, R. Spezia, A. Di Nola, and A. Amadei, A first priciples method
to model perturbed electronic wavefunctions: the effect of an external
electric field. Chem. Phys. Lett., 344:374–380, 2001.
D. B. Kokh, S. Corni, P. J. Winn, M. Hoefling, K. E. Gottschalk, R. C.
Wade, ProMetCS: An Atomistic Force Field for Modeling Protein-Metal
Surface Interactions in a Continuum Aquesous Solvent. J. Chem. Theory
Comput., 6, 1753–1768 (2010).
L. Zanetti-Polzi, I. Daidone, C. A. Bortolotti, and S. Corni, Surface
packing determines the redox potential shift of cytochrome c adsorbed
on gold. Journal of the American Chemical Society, 136:12929–12937,
R. R. Gabdoulline, R. C. Wade, Simulation of the DiffusionalAssociation
of Barnase and Barstar. Biophys. J., 72, 1917–1929 (1997).
Computational Strategies for Protein-Surface 23
www.h-its.org/mcm
D. van der Spoel, E. Lindahl, B. Hess, G. Groenhof, A. E. Mark,
Berendsen, H. J. C, GROMACS: Fast, Flexible, and Free. J. Comp.
Chem., 26, 1701–1718 (2005).
C. Oostenbrink, A. Villa, A. E. Mark, W. F. van Gunsteren, A biomolec-
ular force field based on the free enthalpy of hydration and solvation: the
GROMOS force-field parameter sets 53A5 and 53A6. J. Comp. Chem.,
, 1656–1676 (2004).
N. Spackova, I. Berger, M. Egli, J. Sponer, Molecular Dynamics of
Hemiprotonated Intercalated Four-Stranded i-DNA: Stable Stable Tra-
jectories on a Nanosecond Scale J. Am. Chem. Soc., 120, 6147–6151
(1998).
L. B. Wright, M. P. Rodgera, T. R. Walsh, Aqueous citrate: a first-
principles and force-field molecular dynamics study RSC Adv., 3,
–16409 (2013).
J.-W. Park, J. S. Shumaker-Parry, Structural Study of Citrate Layers on
Gold Nanoparticles: Role of Intermolecular Interactions in Stabilizing
Nanoparticles. J. Am. Chem. Soc., 136, 1907–1921 (2014).
J. Kunze, I. Burgess, R. Nichols, I. Buess-Herman, J. Lipkowski,
Electrochemical Evaluation of Citrate Adsorption on Au(111) and the
Stability of Citrate-Reduced Gold Colloids. J. Electroanal. Chem., 599,
–159 (2007).
http://biophysics.cs.vt.edu/H++
J. L. Elechiguerra, J. Reyes-Gasga, M. J. Yacaman, The Role of Twinning
in Shape Evolution of Anisotropic Noble Metal Nanostructures J. Mater.
Chem., 16, 3906–3919 (2006).
Y. Lin, G. Pan, G.-J. Su, X.-H. Fang, L.-J. Wan, C.-L. Bai, Study of
CitrateAdsorbed on theAu(111) Surface by Scanning Probe Microscopy.
Langmuir, 19, 10000–10003 (2003).
http://projects.villa-bosch.de/mcmsoft/sda/6.00/
R. R. Gabdoulline, R. C. Wade, Effective Charges for Macromolecules
in Solvent. J. Phys. Chem., 100, 3868–3878 (1996).
G. Brancolini, A. Corazza, M. Vuano, F. Fogolari, M. C. Mimmi, V.
Bellotti, M. Stoppini, S. Corni, G. Esposito, to be submitted 2014
L. Zanetti-Polzi, A. Amadei, M. Aschi, and I. Daidone, Insight into the
ir-spectra/structure relationship in amyloid fibrils: a theoretical study on
a prion peptide. J. Am. Chem. Soc., 133(30):11414–11417 (2011).
A. Amadei, I. Daidone, and M. Aschi, A general theoretical model for
electron transfer reactions in complex systems. Phys. Chem. Chem.
Phys., 14:1360–13770 (2012).
G. Brancolini et al.
I. Daidone,A.Amadei, F. Zaccanti, M. Borsari, and C.A. Bortolotti, How
the reorganization free energy affects the reduction potential of struc-
turally homologous cytochromes. The Journal of Physical Chemistry
Letters, 5(9):1534–1540 (2014).
R. Spezia, M. Aschi, A. Di Nola, and A. Amadei, Extension of the
perturbed matrix method: application to a water molecule. Chem. Phys.
Lett., 365:450–456 (2002).
A. Amadei, M. D’Abramo, C. Zazza, and M. Aschi, Electronic properties
of formaldehyde: a theoretical study. Chem. Phys. Lett., 381:187–193
(2003).
A. Amadei, F. Marinelli, M. D’Abramo, M. D’Alessandro, M. Anselmi,
A. Di Nola, and M.Aschi, Theoretical modeling of vibro-electronic quan-
tum states in complex molecular systems: solvated carbon monoxide, a
test case. J. Chem. Phys., 122:124506 (2005).
A. Amadei, M. D’Alessandro, M. D’Abramo, and M. Aschi, Theoretical
characterization of electronic states in interacting chemical systems.
J. Chem Phys., 130:08410–08415 (2009).
J. Gao and D. G. Truhlar, Quantum mechanical methods for enzyme
kinetics. Ann. Rev. Phys. Chem., 53:467–505 (2002).
T. Vreven and K. Morokuma, Chapter 3 hybrid methods: ONIOM
(QM:MM) and QM/MM. Ann. Rep. Comp. Chem., 2:35–51 (2006).
H. M. Senn and W. Thiel, QM/MM studies of enzymes. Curr. Opin.
Chem. Biol., 11:182–187 (2007).
A. Amadei, M. D’Alessandro, and M. Aschi, Statistical mechanical
modeling of chemical reactions in complex systems: the reaction free
energy surface. J. Phys. Chem. B, 108:16250–16254 (2004).
I. Muegge, P. X. Qi, A. J. Wand, Z. T. Chu, and A. Warshel, The
reorganization energy of cytochrome c revisited. J. Phys. Chem. B,
:825–836 (1997).
L. B. Sagle, J. Zimmermann, S. Matsuda, P. E. Dawson, and F. E.
Romesberg, Redox-coupled dynamics and folding in cytochrome c.
J. Am. Chem. Soc., 1128(24):7909–7915 (2006).
H. A. Heering, F. G. M. Wiertz, C. Dekker, and S. deVries, Direct immo-
bilization of native yeast iso-1 cytochrome c on bare gold: fast electron
relay to redox enzymes and zeptomole protein-film voltammetry. J. Am.
Chem. Soc., 126:11103–11112 (2004).
C. A. Bortolotti, G. Battistuzzi, M. Borsari, P. Facci, A. Ranieri, and
M. Sola, The redox chemistry of the covalently immobilized native
and low-pH forms of yeast iso-1-cytochrome c. J. Am. Chem. Soc.,
:5444–5451 (2006).
Computational Strategies for Protein-Surface 25
C. A. Bortolotti, A. Amadei, M. Aschi, M. Borsari, S. Corni, M. Sola, and
I. Daidone, The reversible opening of water channels in cytochrome c
modulates the heme iron reduction potential. J. Am. Chem. Soc.,
:13670–13678 (2012)