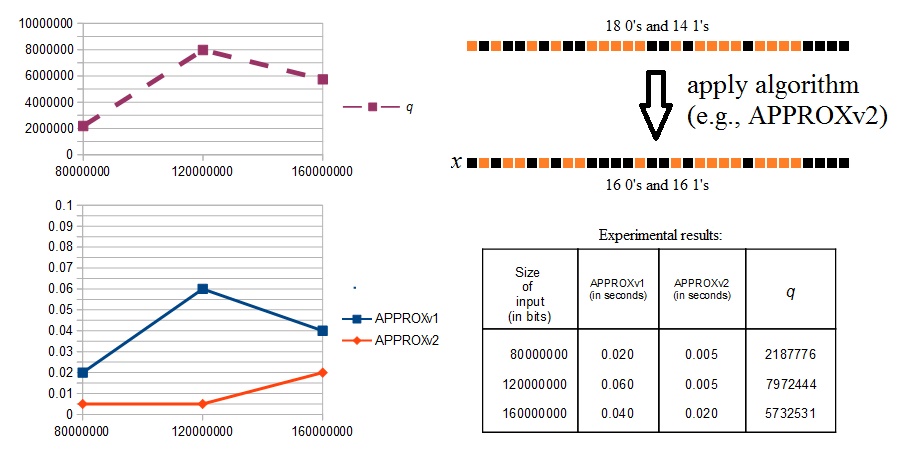
Very Fast Algorithms for Eliminating the Diffraction Effects in Protein-Based Volumetric Memories
DOI:
https://doi.org/10.13052/jsame2245-4551.213Keywords:
compression, biological nanotechnology, optimizationAbstract
One of the current research directions in biological nanotechnology is the use
of bacteriorhodopsin in the fabrication of protein-based volumetric memories.
Bacteriorhodopsin, with its unique light-activated photocycle, nanoscale size,
cyclicity (>107), and natural resistance to harsh environmental conditions,
provides for protein-based volumetric memories that have a comparative
advantage over magnetic and optical data storage devices. The construction
of protein-based volumetric memories has been, however, severely limited by
fundamental issues that exist with such devices, such as unwanted diffraction
effects. In this paper, we propose some optimizations that can be applied to one
of the previously proposed algorithms for eliminating the diffraction effects.
Downloads
References
R. R. Birge, Photophysics and molecular electronic applications of the
rhodopsins, Annual Review of Physical Chemistry, vol. 41, pp. 683–733
(1990)
R. R. Birge, N. B. Gillespie, E. W. Izaguirre, A. Kusnetzow, A. F.
Lawrence, D. Singh, Q. W. Song, E. Schmidt, J.A. Stuart, S. Seetharaman
and K. J. Wise, Biomolecular electronics: protein-based associative
processors and volumetric memories, Journal of Physical Chemistry B,
vol. 103, pp. 10746–10766 (1999)
N. A. Hampp, Bacteriorhodopsin: mutating a biomaterial into an opto-
electronic material, Applied Microbiology and Biotechnology, vol. 53,
pp. 633–639 (2000)
Very Fast Algorithms for Eliminating the Diffraction Effects 51
J. R. Hillebrecht, J. F. Koscielecki, K. J. Wise, D. L. Marcy, W. Tetley,
R. Rangarajan, J. Sullivan, M. Brideau, M. P. Krebs, J. A. Stuart and
R. R. Birge, Optimization of protein-based volumetric optical memories
and associative processors by using directed evolution, NanoBiotech-
nology, vol. 1, pp. 141–151 (2005)
D. Marcy, W. Tetley, J. Stuart and R. Birge, Three-dimensional data
storage using the photochromic protein bacteriorhodopsin, in Proc. of
the 22nd Annual International Conference of the IEEE Engineering in
Medicine and Biology Society (EMBC’00), pp. 1003–1006
D. Oesterhelt, C. Br ̈auchle and N. Hampp, Bacteriorhodopsin: a bio-
logical material for information processing, Quarterly Reviews of
Biophysics, vol. 24, pp. 425–478 (1991)
S. Rajasekaran, V. Kumar, S. Sahni and R. Birge, Efficient algorithms for
protein-based associative processors and volumetric memories, in Proc.
of IEEE NANO 2008, pp. 397–400
S. Rajasekaran, V. Kundeti, R. Birge, V. Kumar and S. Sahni, Efficient
algorithms for computing with protein-based volumetric memory pro-
cessors, IEEE Transactions on Nanotechnology, vol. 10, pp. 881–890
(2010)
D. Trinc ̆a and S. Rajasekaran, Coping with diffraction effects in protein-
based computing through a specialized approximation algorithm with
constant overhead, in Proc. of IEEE NANO 2010, pp. 802–805
D. Trinc ̆a and S. Rajasekaran, Specialized compression for coping with
diffraction effects in protein-based volumetric memories: solving some
challenging instances, Journal of Nanoelectronics and Optoelectronics,
vol. 5, pp. 290–294 (2010)
D. Trinc ̆a and S. Rajasekaran, Optimizing the APPROXv1 algorithm for
coping with diffraction effects in protein-based volumetric memories, in
Technical Summaries of SPIE Optical Systems Design, abstract 8167A-
(at page 14), 2011
D. Trinc ̆a and S. Rajasekaran, Fast Algorithms for Coping with Diffrac-
tion Effects in Protein-based Volumetric Memories (Design and Imple-
mentation), Journal of Computational and Theoretical Nanoscience,
vol. 10, pp. 894–897 (2013)