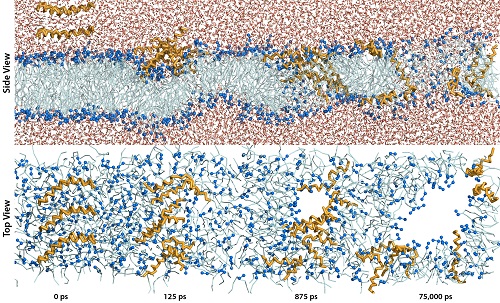
An Investigation of the Interaction between Melittin and a Model Lipid Bilayer
DOI:
https://doi.org/10.13052/jsame2245-4551.214Keywords:
Antimicrobial peptides, melittin, molecular dynamic simulations, self-assembled melittin/lipid bilayer poreAbstract
The recent emergence of multi-drug resistant bacteria has rendered many
common antibiotics ineffective and therefor novel drugs are needed. Antimi-
crobial peptides have proven applicable in this aspect as they can form
self-assembled pore structures in the lipid bilayer of bacterial cells. The
exact pore formation is shrouded in controversy and largely unknown as it is
difficult to determine empirically. The development of coarse grain force fields
in molecular dynamic simulations enables longer timescales and extended
simulation systems. This allows for the simulation of lipid bilayer self-
assembly and bilayer-antimicrobial peptide interactions. The present work
focuses on the antimicrobial peptide, melittin, induced pore formation and is
aimed towards determining the mechanism and pore structure
Downloads
References
D. Shingadia and V. Novelli. Diagnosis and treatment of tuberculosis in
children. Lancet Infect Dis, 3: 624–632 (2003).
M. Hassan, M. Kjos, I.F. Nes, D.B. Diep and F. Lotfipour. Natural antimi-
crobial peptides from bacteria: characteristics and potential applications
to fight against antibiotic resistance. Journal of Applied Microbiology,
: 723–736 (2012).
H. G. Boman. Antibacterial peptides: basic facts and emerging concepts.
Journal of Internal Medicine, 254(3): 197–215 (2003).
Kim A. Brogden Antimicrobial peptides: pore formers or metabolic
inhibitors in bacteria? Nature Reviews Microbiology, 3: 238–250 (2005).
MR. Yeaman and NY. Yount Mechanisms of antimicrobial peptide action
and resistance. Pharmacological Reviews, 55(1): 27–55 (2003).
Kristopher Hall, Tzong-Hsien Lee and Marie-Isabel Aguilar The role of
electrostatic interactions in the membrane binding of melittin. Journal
of Molecular Recognition, 24(1): 108–18 (2011).
H. Raghuraman and Amitabha Chattopadhyay. Melittin: a Membrane-
active Peptide with Diverse Functions. Biosience Reports, 27:189–223
(2007).
Y. -H. Lam, S. R. Wassall, C. J. Morton, R. Smith and F. Separovic. Solid-
State NMR Structure Determination of Melittin in a Lipid Environment.
Biophysical Journal, 81: 2752–2761 (2001).
Katsumi Matsuzaki, Shuji Yoneyama, and Koichiro Miyajima. Pore For-
mation and Translocation of Melittin. Biophysical Journal, 73: 831–838
(1997).
An Investigation of the Interaction between Melittin and a Model Lipid Bilayer 73
M. T. Tosteson and D. C. Tosteson. The sting. Melittin forms channels
in lipid bilayers. Biophysical Journal, 36: 109–116 (1981).
H. Vogel and F. J ̈ahnig. The structure of melittin in membranes.
Biophysical Journal, 50: 573–582 (1986).
Durba Sengupta, Hari Leontiadou, Alan E. Mark and Siewert-Jan
Marrink. Toroidal pores formed by antimicrobial peptides show sig-
nificant disorder. Biochimica et Biophysica Acta, 1778: 2308–2317
(2008).
Michal Sharon, Ziv Oren, Yechiel Shai, and Jacob Anglister. 2D-NMR
and ATR-FTIR Study of the Structure of a Cell-Selective Diastereomer
of Melittin and Its Orientation in Phospholipids. Biochemistry, 38:
–15316 (1999).
Ziv Oren and Yechiel Shai. Mode of action of linear amphipathic a-helical
antimicrobial peptides. Peptide Science, 47: 451–463 (1998).
F. Inagaki, K. Shimada, K. Kawaguchi, M. Hirano, I. Terasawa, T. Ikura
and N. Go. Structure of melittin bound to perdeuterated dodecylphos-
phocholine micelles as studied by two-dimensional NMR and distance
geometry calculations. Biochemistry, 28: 5985-5991 (1989).
R. Smith, F. Separovic, T. J. Milne, A. Whittaker, F. M. Bennett, B.
A. Cornell and A. Makriyannis. Structure and orientation of the pore-
forming peptide, melittin, in lipid bilayers. Journal of molecular biology,
: 456-466 (1994).
Akihiko Okada, Kaori Wakamatsu, Tatsuo Miyazawa and Tsutomu
Higashijima. Vesicle-Bound Conformation of Melittin: Transferred
Nuclear Overhauser Enhancement Analysis in the Presence of Perdeuter-
ated Phosphatidylcholine Vesicles. Biochemistry, 33: 9438–9446
(1994).
Shuichi Toraya, Katsuyuki Nishimura, and Akira Naito. Dynamic Struc-
ture of Vesicle-Bound Melittin in a Variety of Lipid Chain Lengths by
Solid-State NMR. Biophysical Journal, 87: 3323–3335 (2004).
Geert van den Bogaart, Jacek T. Mika, Victor Krasnikov and Bert
Poolman. The Lipid Dependence of Melittin Action Investigated by
Dual-Color Fluorescence Burst Analysis. Biophysical Journal, 93(1):
–163 (2007).
Xiaoyun Chen, Jie Wang, Andrew P. Boughton, Cornelius B. Kristalyn,
and Zhan Chen. Multiple Orientation of Melittin inside a Single Lipid
Bilayer Determined by Combined Vibrational Spectroscopic Studies.
J. Am. Chem. Soc., 129: 1420–1427 (2007).
M. Slyngborg et al.
H. Raghuraman and Amitabha Chattopadhyay. Orientation and Dynam-
ics of Melittin in Membranes of Varying Composition Utilizing NBD
Fluorescence. Biophysical Journal, 92: 1271–1283 (2007).
Jung-Hsin Lin and A. Baumgaertner. Stability of a Melittin Pore in a
Lipid Bilayer: A Molecular Dynamics Study. Biophysical Journal, 78:
–1724 (2000).
S. Rex. Pore formation induced by the peptide melittin in different lipid
vesicle membranes. Biophysical Chemistry, 58: 75-85 (1996).
A. S. Ladokhin, M. E. Selsted, and S. H. White. Sizing membrane pores
in lipid vesicles by leakage of co-encapsulated markers: pore formation
by melittin. Biophysical Journal, 72: 1762-1766 (1997).
Maja Mihajlovic and Themis Lazaridis. Antimicrobial Peptides in
Toroidal and Cylindrical Pores. Biochim Biophys Acta., 1798(8):
–1493 (2010).
Alice Gl ̈attli, Indira Chandrasekhar and Wilfred F. van. Gunsteren. A
molecular dynamics study of the bee venom melittin in aqueous solution,
in methanol, and inserted in a phospholipid bilayer. Eur Biophys Journal,
: 255–267 (2006).
Siewert J. Marrink, H. Jelger Risselada, Serge Yefimov, D. Peter Tiele-
man and Alex H. de Vries. The MARTINI Force Field: Coarse Grained
Model for Biomolecular Simulations. J. Phys. Chem. B, 111: 7812–7824
(2007).
Wolfgang Kabsch and Chris Sander. Dictionary of protein secondary
structure: pattern recognition of hydrogen-bonded and geometrical
features. Nucleic Acids Research, 22: 2577–2637 (1983).
Y. Wu, K. He, S. J. Ludtke and H. W. Huang. X-ray diffraction study
of lipid bilayer membrane interacting with amphiphilic helical peptides:
diphytanoyl phosphatidylcholine with alamethicin at low concentrations.
Biophys. Journal, 68: 2361–2369 (1995).
S. Ludtke, K. He, and H. W. Huang. Membrane thinning caused by
magainin 2. Biochemistry, 34: 16764–16769 (1995).
W. T. Heller, A. J. Waring, R. I. Lehrer, T. A. Harroun, T. M. Weiss,
L. Yang and H. W. Huang. Membrane thinning effect of the b-sheet
antimicrobial protegrin. Biochemistry, 39: 139–145 (2000).
A. Naito, T. Nagao, K. Norisada, T. Mizuno, S. Tuzi and H.
Saitˆo. Difference between Magainin-2 and Melittin Assemblies in
An Investigation of the Interaction between Melittin and a Model Lipid Bilayer 75
Phosphatidylcholine Bilayers: Results from Coarse-Grained Simula-
tions. The Journal of Physical Chemistry B, 116, 3021–3030 (2012).
Kolattukudy P. Santo and Max L. Berkowitz. Conformation and dynam-
ics of melittin bound to magnetically oriented lipid bilayers by solid-state
P- and 13 C-NMR spectroscopy. Biophys. Journal, 78: 24052417
(2000).
M. Zasloff. Antimicrobial peptides of multicellular organisms. Nature,
, 389–395 (2002)